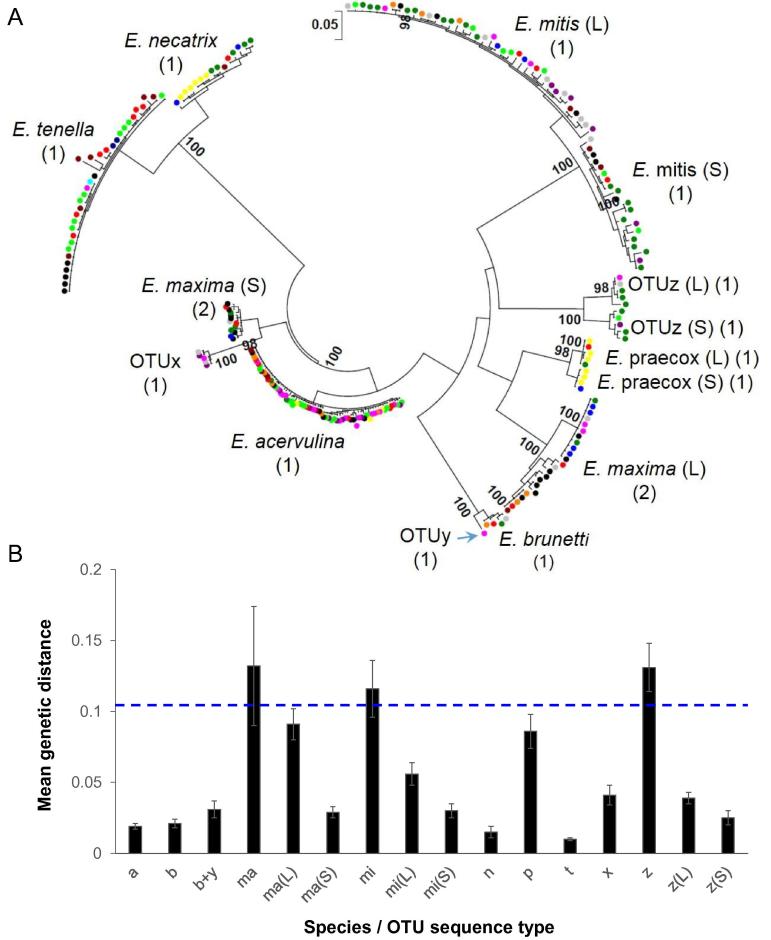
Fig. 2.
Eimeria spp. and operational taxonomic unit (OTU) internal transcribed spacer sequence (ITS) 1 and 2 diversity. (A) Tamura-Nei model Maximum Likelihood (ML) phylogeny of ITS1-5.8S-ITS2 sequences derived with 1000 bootstrap replication from samples collected in Asia, Africa, Europe and the Americas (GenBank accession numbers LN609768–LN609975). The number and origin of sequences used are shown in Table 1. Coloured spots indicate the country of origin for each sequence (Asia: China (blue), India (dark blue), Japan, (light blue). Europe: countries pooled (red). North Africa: Egypt (orange), Libya (dark red). Sub-Saharan Africa: Ghana (purple), Nigeria (pink), Tanzania (dark green), Uganda (green), Zambia (yellow). Americas: USA (black), Venezuela (grey)). L, long sequence form; S, short sequence form. Numbers shown in parentheses indicate the number of putative species partitioned per recognised species, sequence form or OTU by Bayesian species delimitation. (B) Mean genetic distance within each species and OTU genotype calculated using ML with 1000 bootstrap replications. Species and genotype identifiers are as follows: a, Eimeria acervulina; b, Eimeria brunetti; ma, Eimeria maxima; mi, Eimeria mitis; n, Eimeria necatrix; p, Eimeria praecox; t, Eimeria tenella; x, OTUx; y, OTUy; z, OTUz; L, long sequence form; S, short sequence form. The dotted line indicates the intersect between the combined and the separated long and short sequence forms.