Figure 2.
MAIL, A Localization Motif for Importin β1 mRNA
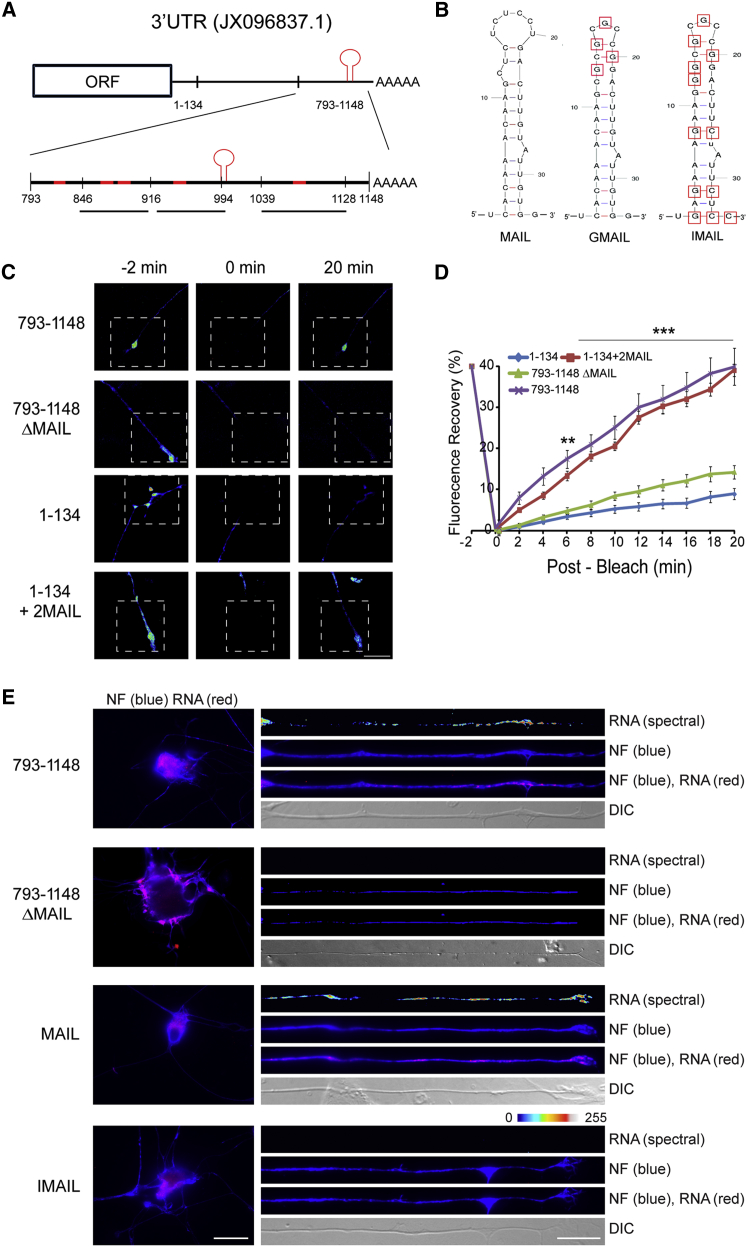
(A) Schematic diagram of segments from the importin β1 3′ UTR (GenBank: JX096837.1) evaluated for axon-localizing activity. Regions predicted to contain stem-loop secondary structures are highlighted in red. The region between 1 and 134 nt encompasses the short form of importin β1 3′ UTR, which is restricted to the cell body. The motif for axonal importin localization (MAIL) is shown as a red stem-loop structure at 991–1,024 nt.
(B) Sequences and schematic structure predictions of the MAIL motif and two derived mutants, GMAIL, with four U-G mutations in the loop region as shown, and IMAIL, which carries the GMAIL mutations together with additional mutations in the stem region, are shown.
(C) Constructs containing deletions or fusions of the MAIL motif as indicated were fused with a destabilized myr-EGFP reporter and transfected to sensory neurons for FRAP analyses, with recovery monitored over 20 min. Representative images from time-lapse sequences before (−2 min) and after photobleaching (0 and 20 min) in the boxed region of interest are shown. For data from additional constructs, see Figure S2A. Scale bar, 25 μm.
(D) Quantification of the FRAP analyses shown in (C). Average recoveries are shown (percentage of pre-bleach levels ± SEM). Anisomycin-treated neurons were exposed to 50 μM inhibitor prior to the imaging sequence. Time points with significant differences in axonal fluorescence compared to that observed in anisomycin-treated cultures are indicated (∗∗∗p < 0.001 and ∗∗p < 0.01, two-way ANOVA). For results with additional deletion constructs and anisomycin controls, see Figure S2B.
(E) In situ hybridization on neurons transfected with the indicated constructs. Exposure-matched images show that only GFP mRNA with the MAIL element localizes into axons (right panel), while all reporter mRNAs are clearly expressed in corresponding cell body images (left panel). Scale bars, 25 μm (cell body) and 10 μm (axons). See also Figure S2C.
See also Figure S2.