Figure 3.
Binding of All EJC Components Is Highly Enriched to Transcripts Coding for RNA Processing Proteins
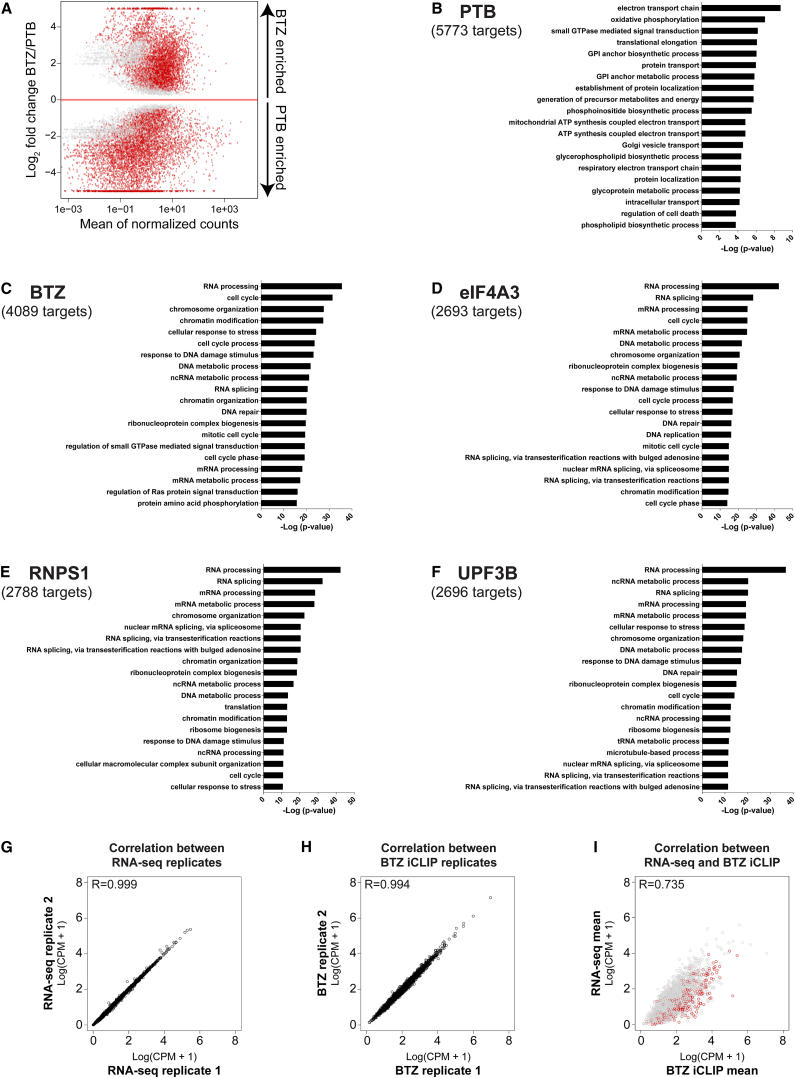
(A–F) The red dots show significant mRNA targets either up- or downregulated after differential analysis using edgeR controlled by the Benjamini-Hochberg procedure with an FDR <0.05. The targets below the red line provided a higher PTB signal, whereas targets above the red line exhibited a higher BTZ signal (A). The targets below the red line were used for GO enrichment analysis of PTB (B). All EJC iCLIP data sets were compared to PTB, and the targets with a log2 fold change >0 (above red line) were analyzed for GO enrichment for BTZ (C), eIF4A3 (D), RNPS1 (E), and UPF3B (F).
(G–I) Enriched EJC occupancy does not correlate with mRNA abundance. These plots display the correlation of iCLIP and RNA-seq data using the 2,194 common targets that were significantly enriched in BTZ iCLIP in both differential analyses (compared to PTB and GFP).
(G and H) The RNA-seq (G) and BTZ iCLIP (H) replicates were highly reproducible.
(I) Relationship between the mean count of all three RNA-seq and BTZ iCLIP libraries. The red dots highlight mRNA targets for BTZ with a log2 fold change >3 compared to PTB in the differential analysis using edgeR and thus indicate mRNAs that are particularly strongly bound by BTZ. The specificity of these highly occupied mRNAs is demonstrated by the finding that these mRNAs were distributed across transcripts with high and low expression levels as measured by RNA-seq (CPM and R = Pearson's correlation coefficient).
See also Figure S3.