Figure 5.
Binding of EJC Components Is Highly Enriched at Alternatively Spliced Exons in Transcripts with High EJC Occupancy and Enables Detection of Low-Abundance NMD-Sensitive mRNA Isoforms
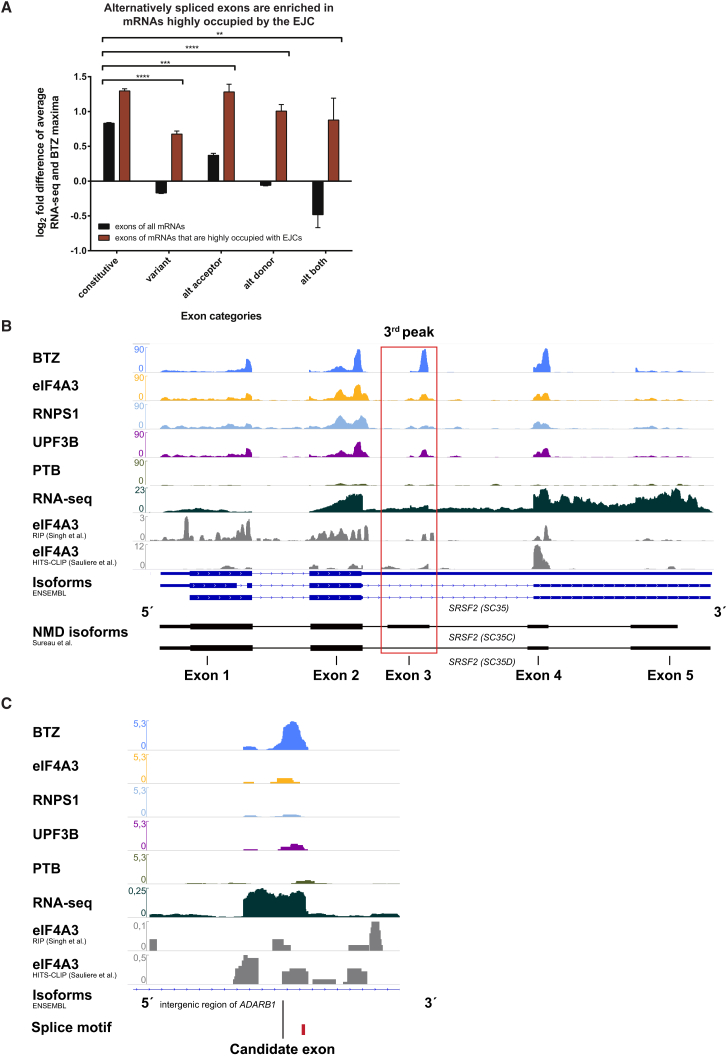
(A) We used the RNA-seq and BTZ maxima as shown in Figure 4 (see also Table S4) to calculate a log2 fold difference for both all mRNAs and mRNAs that are highly occupied by EJCs. The iCLIP/RNA-seq ratio is enriched for alternatively spliced exons in RNAs that are highly occupied by EJCs (see Figures 3G–3I) compared to all mRNAs.
(B) Genome browser view of SRSF2 (SC35) gene reveals EJC iCLIP peaks on exons corresponding to NMD-sensitive SC35C and SC35D mRNA isoforms. The NMD-insensitive SC35WT isoform is displayed in the Ensembl genes track as the upper isoform. The red box highlights the variant exon 3.
(C) Candidate exon in the intronic region of the mRNA ADARB1. The track range displays CPM and was adjusted to the highest iCLIP signal obtained in the iCLIP libraries of this study in each genome browser view. The signals of the RNA-seq and literature data were not adjusted. The literature data were obtained from RIP (Singh et al., 2012) and HITS-CLIP (Saulière et al., 2012) of eIF4A3.
See also Figures S4 and S6.