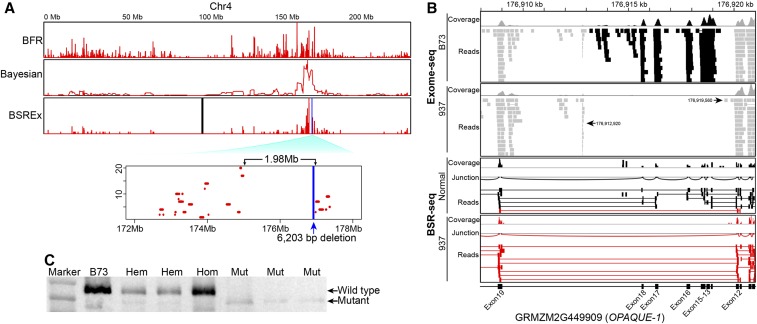
Figure 4.
Identification of causative deletion candidate in mutant 937 by combining BSR-seq and exome-seq. (A) The current BSR-seq analysis and two other BSR-seq methods, Bayesian (Liu et al. 2012) and BFR (Ramirez-Gonzalez et al. 2015), all mapped the causative gene to chromosome 4 in mutant 937. Enlargement of a sharp linkage peak (red), 172–178 Mb, and a gap (∼1.98 Mb) covering the 6203 bp deletion (blue) spanning six exons of O1 gene inside the linkage peak were shown. Black bar: centromere. Y axis scale for positive SNPs/indels ranges from 0 to 20. (B) The alignments of exome-seq reads (gray) around the causative deletion, including six exons (#13–#18, in black), are shown in the coverage and read windows. The alignment of BSR-seq reads showed two transcript variants, one for normal type (black), and the other (red) for mutant 937 without six exons of o1 gene. (C) Western blot shows two protein variants with different lengths: wild-type and mutant truncated proteins. Hem, hemizygous-type kernel with both wild-type and mutant truncated proteins; Hom, homozygous wild-type kernel with only wild-type proteins; Mut, mutant-type kernel with only mutant truncated proteins.