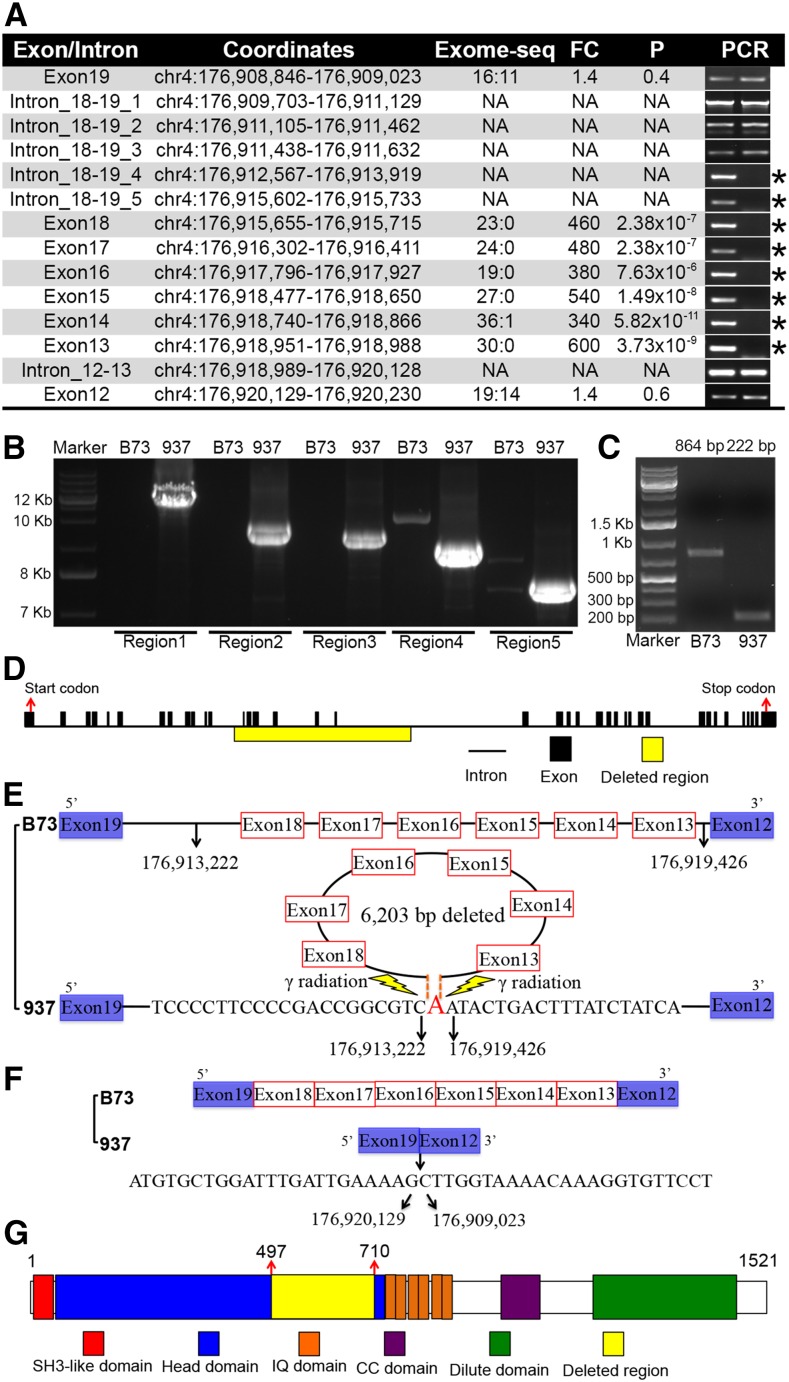
Figure 5.
Genomic PCR and clone verification of deletion in O1 in mutant 937. (A) Exon/Intron column is for the region where primers were designed. Mapped read depth coverage in exome-seq was normalized to compare wild-type B73 and mutant 937 (B73:937). Fold-changes (FC) of read depth coverage between B73 and mutant 937 and P-values were used to search for deleted exons. The verified deletions are noted with an asterisk. (B) Five long PCRs were conducted to cover the junction site, and PCR product sizes are 7584 bp, 8331 bp, 8713 bp, 9046 bp, and 11,327 bp for Region 1–5. No bands in B73 result from genomic regions too long for PCR. (C) cDNA was synthesized and RT-PCR was conducted to compare B73 and 937 and showed an ∼600 bp difference. Clone sequencing confirmed the PCR product sizes are 864 bp in B73 and 222 bp in 937. (D) The causative deletion covered six exons inside the total 39 exons of O1 gene. (E) Clone sequencing of genomic PCR products in 937 confirmed the 6203 bp deletion inside O1 gene and the two break points at 176,913,222 and 176,919,426 based on maize genome v3.25. (F) cDNA clone sequencing identified the transcript variant without the six deleted exons, and exons #12 and #19 were linked directly. The O1 gene resides in the negative strand, and was shown in coordinates for the positive strand from 5′ end to 3′ end. (G) Protein domains in O1. The deletion from position 497 to 710 was in the head domain. Protein domain positions were from Wang et al. (2012).