Figure 1.
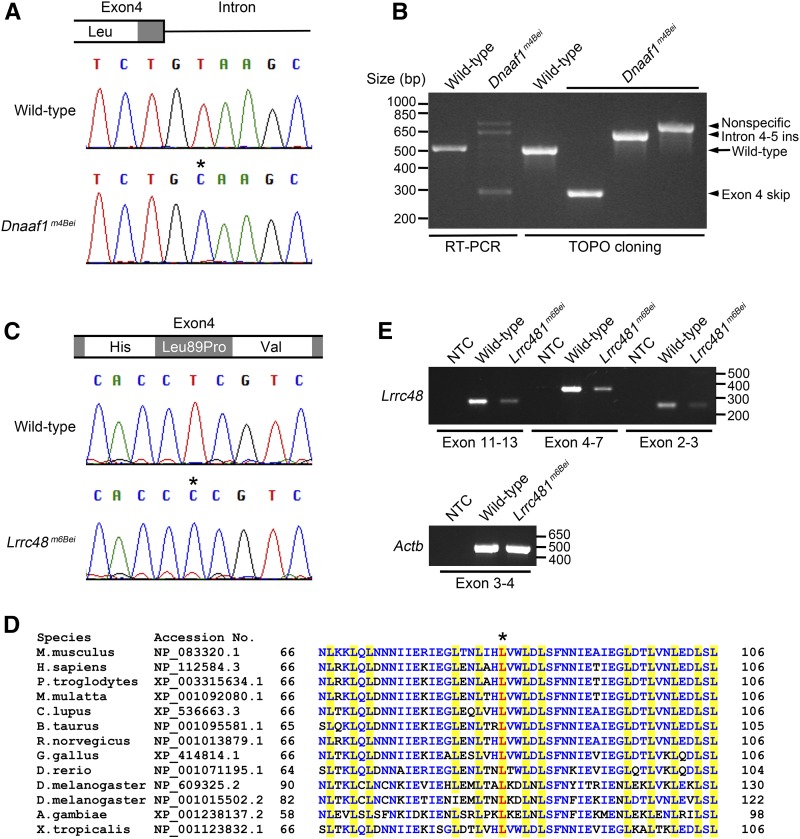
Mutations in Dnaaf1 and Lrrc48. (A) A chromatogram from Sanger sequencing showing a T > C mutation (asterisk) at the junction of exon 4 and intron. (B) RT-PCR result showing amplification of a fragment corresponding to exon 3–6 of Dnaaf1 (lanes 1 and 2). Multiple bands with incorrect sizes were detected in Dnaaf1m4Bei, indicating a splicing defect. Cloned RT-PCR products revealed abnormal transcripts with exon 4 skipping (lane 4, insert size 279 bp) or intron 4–5 insertion (lane 5, insert size 617 bp). The largest band (nonspecific) was not a Dnaaf1 transcript. (C) A chromatogram from Sanger sequencing showing a T > C mutation (asterisk) causing an amino acid change (Leu89Pro). (D) Multiple protein sequence alignment adapted from NCBI HomoloGene search result, including conserved leucine-rich repeat domains near the mutation. Leucine residues in the domain are highlighted in yellow. The leucine residue substituted in Lrrc48m6Bei mutants is shown in red (asterisk). Other conserved residues are marked in blue. (E) RT-PCR results using three different primer sets spanning the Lrrc48 transcript. All amplified weaker bands from the mutant. Actb (Beta-actin) was used as a control. NCBI, National Center for Biotechnology Information; NTC, no template control; RT-PCR, reverse transcription polymerase chain reaction.