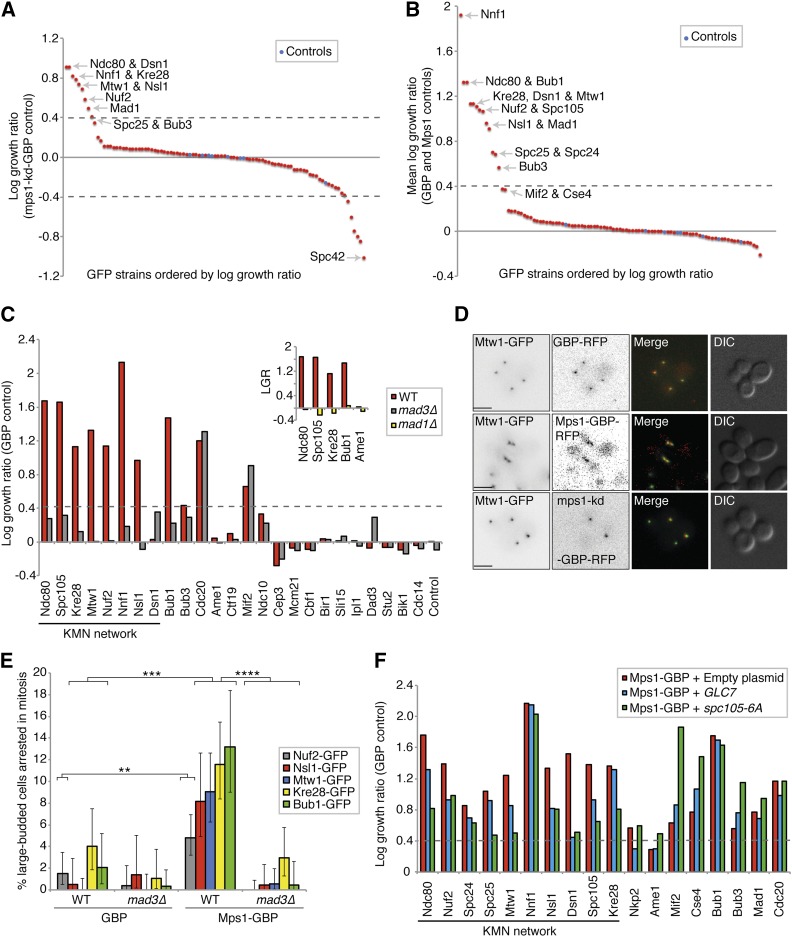
Figure 5.
Mps1 recruitment to the KMN network results in a growth defect. (A) Mps1-GBP and a kinase-dead control (mps1-kd-GBP) were transferred into the 88 kinetochore-related GFP strains. A number of GFP strains show growth defects, including most members of the KMN network of proteins. The strains labeled ‘Controls’ in blue are an untagged BY4741 strain (File S4). (B) We also compared Mps1-GBP with two other controls (Mps1 or GBP alone), and all of the KMN network of proteins produce a growth defect (File S4). (C) We deleted the MAD3 gene in 25 of these GFP strains and repeated the assay with Mps1-GBP compared with GBP alone. (Inset) We also deleted MAD1 in several GFP strains. The KMN-Mps1 (and Bub1) SPIs are all suppressed by deletion of MAD3 and MAD1 (except Dsn1). LGR, log growth ratio. (D) Fluorescence image analysis confirms the colocalization of Mps1 with the KMN protein Mtw1; scale bars are 5 µm. (E) Cell cycle progression analysis of Mps1 SPIs. Colonies from the SPI assay were grown in culture overnight, imaged the following day with fluorescent microscopy, and cells counted (ranging from 141 to 560 cells for each condition). The graph shows the percentage of large-budded cells with two GFP-kinetochore foci in close proximity. Large-budded cells with separated kinetochore foci were discounted. Statistical analysis was done using Fishers exact test; **** p < 0.0001. *** p < 0.001, ** p < 0.01. Error bars indicate 95% binomial C.I. (F) Mps1-GBP and the GBP control were transferred into the 88 kinetochore-related GFP strains. In addition, three separate plasmids were also transferred simultaneously into these GFP strains, one empty, one with pCUP1-GLC7, and the third with pGALS-spc105-6A. The graph shows a selection of the resulting data, and shows that both increased expression of GLC7 (blue bars), and overexpression of spc105-6A (green bars), suppress the Mps1 SPI growth phenotype specifically at the KMN network compared to the empty plasmid (red bars).