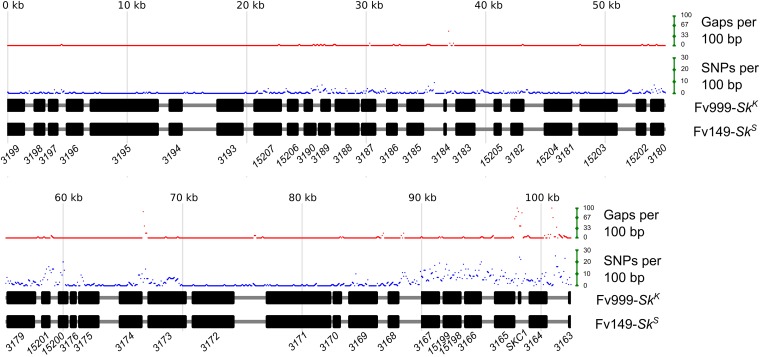
Figure 4.
Strain Fv999-SkK carries a unique gene within a hyper-variable region of the Sk region. The Sk regions from Fv999-SkK (GenBank, KU963213) and Fv149-SkS (GenBank, CM000582.1, positions 665,669 to 767,411) were imported into BioEdit (Version 7.2.5) (Hall 1999) and aligned with Clustal W. Custom Perl scripts were used to examine base mismatches and gap positions, and to generate the diagram. The total number of mismatches (SNPs) between the two sequences was calculated for each 100-base nonoverlapping window of the alignment. The total number of gaps was also calculated for each 100-base nonoverlapping window. A gap position was not considered an SNP. By this definition, a window with 100 gaps cannot have SNPs. Black rectangles represent the coding regions of predicted genes. Gene names have been abbreviated according to their identification tags in the Fv149-SkS annotation. For example: FVEG_03199 was shortened to 3199. A striking feature of the alignment is the presence of three hyper-variable intervals; spanning FVEG_03180 to FVEG_03175 , FVEG_03174 to FVEG_03173 , and FVEG_03167 to the right border of the Sk region. A second striking feature is the presence of a unique gene, SKC1, in Fv999-SkK only.