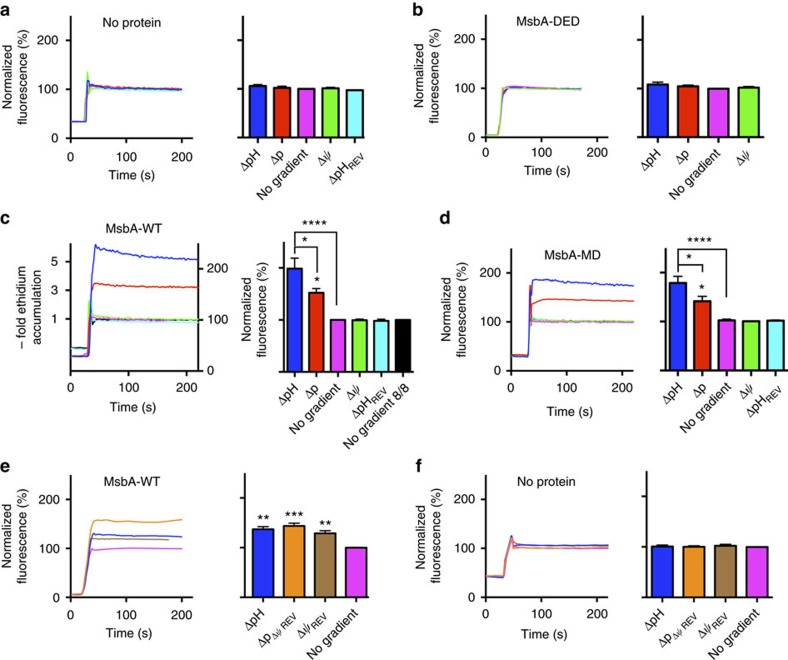
Figure 4. Ethidium transport in proteoliposomes.
(a–d) Ethidium transport in DNA-loaded empty liposomes (a) or proteoliposomes containing the MsbA-DED triple mutant (b), MsbA-WT (c) or MsbA-MD (d) with imposed ΔpH (pHin 6.8/pHout 8.0), Δψ (interior positive), proton-motive force (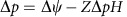 in which Z equals ∼58 mV at 20 °C), ΔpHREV (pHin 8.0/pHout 6.8), or in the absence of ion gradients (pHin 6.8/pHout 6.8, termed No gradient and pHin 8.0/pHout 8.0, No gradient 8/8). No gradient 8/8 for (a,b,d) was very close to the No gradient control, and is not shown for clarity of presentation. The 5-fold accumulation of ethidium by MsbA-WT is indicated in the fluorescence versus time graph in (c). (e,f) Effect of the imposition of a reversed ΔψREV (interior negative) without or with the ΔpH (interior acidic) (
in which Z equals ∼58 mV at 20 °C), ΔpHREV (pHin 8.0/pHout 6.8), or in the absence of ion gradients (pHin 6.8/pHout 6.8, termed No gradient and pHin 8.0/pHout 8.0, No gradient 8/8). No gradient 8/8 for (a,b,d) was very close to the No gradient control, and is not shown for clarity of presentation. The 5-fold accumulation of ethidium by MsbA-WT is indicated in the fluorescence versus time graph in (c). (e,f) Effect of the imposition of a reversed ΔψREV (interior negative) without or with the ΔpH (interior acidic) (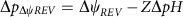 ) on ethidium transport in proteoliposomes containing MsbA-WT (e) or empty liposomes (f). Data represent observations in three or more independent experiments with independently prepared batches of proteoliposomes. Values in histograms show significance of fluorescence levels at steady-state, and are expressed as mean±s.e.m. (one-way analysis of variance; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001).
) on ethidium transport in proteoliposomes containing MsbA-WT (e) or empty liposomes (f). Data represent observations in three or more independent experiments with independently prepared batches of proteoliposomes. Values in histograms show significance of fluorescence levels at steady-state, and are expressed as mean±s.e.m. (one-way analysis of variance; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001).