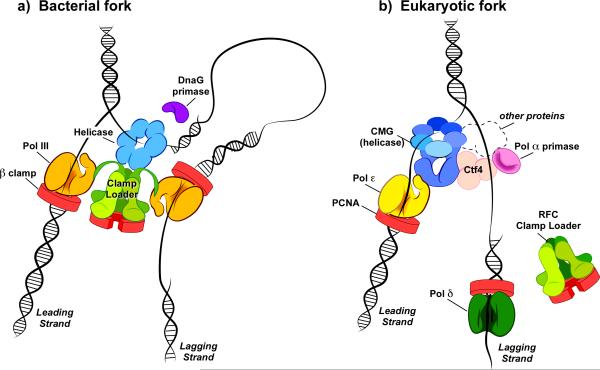
Figure 7.
Distinct replisome machines of bacterial and eukaryotic cells. (a) Replisome of E. coli. The clamp loader organizes the replisome using Cterminal domains that protrude from the three tau subunits of the clamp loader. Each tau subunit binds a separate copy of Pol III core (only two Pol Ill's are shown for clarity but the replisome contains three; see text for details) and each act with a beta clamp to synthesize the leading and lagging strands simultaneously. The tau arms also bind the helicase (DnaB), a homohexamer that encircles the lagging strand. The DnaG primase acts stochastically, binding the helicase periodically to initiate RNA primer synthesis. As RNA primers are extended into Okazaki fragments, DNA loops are formed. (b) Replisome of S. cerevisiae, a model eukaryote. Unlike the bacterial replisome, the organizing unit of the eukaryotic replisome is the CMG helicase, not the clamp loader (RFC). The six Mcm subunits of CMG helicase encircle the leading strand, and the five Cdc45 and GINS accessory factors of CMG form a second channel. CMG binds the Pol epsilon leading strand polymerase. The Ctf4 protein is a trimer that bridges CMG to Pol alpha (i.e. Ctf4 binds both CMG and Pol alpha), and Pol alpha acts as a primase to make hybrid RNA/DNA primers. The lagging strand Pol delta functions with PCNA to extend Okazaki fragments. Tight connection of Pol delta to the replisome is uncertain, and thus may not form DNA loops. Mcm10, Mrc1, Csm3 and Tof1 also travel with the replisome (these subunits are represented by the dashed perimeter) (see color version of this figure at www.informahealthcare.com/bmg).