Abstract
Seizure localization includes neuroimaging like electroencephalogram (EEG), and magnetic resonance imaging (MRI) with limited ability to characterize the epileptogenic network. Temporal clustering analysis (TCA) characterizes epileptogenic network congruent with interictal epileptiform discharges (IED) by clustering together voxels with transient function MRI signals. We generated epileptogenic areas for 12 of 13 epilepsy patients with TCA, congruent with different areas of seizure onset. Resting fMRI scans are non-invasive, and can be acquired quickly (5 min), in patients with different levels of severity and function. Analyzing resting fMRI data using TCA is quick and can be utilized to complement clinical methods to characterize the epileptogenic network.
Keywords: epilepsy, temporal clustering analysis, resting function magnetic resonance imaging, electroencephalogram, seizure foci localization
Introduction
Epilepsy patients that are not responsive to medication require further medical care, thus physicians recommend a resection of potential epileptogenic areas, involving the physical removal of the seizure focus and nearby regions within lobe; successful surgical resections depend on knowing precisely the location of these areas located within the lobe, where the seizure focus resides (Berg 2010, Zijlmans 2007). The current standard of care for seizure localization may at times require assumptions or inferences to be made as it relies on converging evidence from multiple noninvasive and minimally invasive neuroimaging methods. This process is both time consuming and can lead to inconclusive results leading to utilization of more invasive approaches such as subdural grid placement or intradural electrode placement. Below we first outline some of the strengths and drawbacks of existing noninvasive and minimally invasive methods and then describe a relatively new method that involves utilizing resting functional magnetic resonance imaging (fMRI) data to characterize the epileptogenic network.
The initial step in locating epileptogenic areas consists of monitoring interictal brain activity with an electroencephalogram (EEG) recording complemented with a magnetic resonance imaging (MRI) hi-resolution structural brain scan. Recording EEG signals is easy to implement and noninvasive; to determine the seizure focus, clinicians look for abnormal spiking activity that is recorded in a particular electrode. This procedure works well when the seizure focus is located near the scalp and the activity occurs during an ictal state. However, the seizure can rarely be localized during an interictal state (Krakow 2008). EEG is overall limited by low spatial resolution and also by the depth of seizure focus (Khatamian 2011, Lopes 2012).
Structural MRI provides high spatial resolution, is noninvasive and acquisition requires minimal effort from both the clinician and the patient (Zijlmans 2007). Radiologists look for detectable lesions or obvious abnormalities within the brain tissue by examining one slice at a time, a tedious and time consuming task that can result in the neurologist missing an abnormality. Further, often times there are no detectable lesions associated with the region of the brain where the seizure originates (Morgan 2004).
Some studies have explored a novel technique that records functional MRI (fMRI) and EEG simultaneously (Kobayashi 2006, Salek-Haddadi 2002). EEG detects interictal epileptiform discharges, while areas with significant signal intensity differences are detected with fMRI, whose response occurs a few seconds after discharges (Morgan 2010, Zijlmans 2007). Using fMRI and EEG, simultaneously, overcomes some of the individual limitations; however, the simultaneous recordings are not sensitive to activity in deep structures and are usually only accurate with patients that experience frequent interictal epileptiform discharges recorded on the scalp (Lopes 2012, Morgan 2004). In addition, this technique is also very cumbersome, time consuming, and expensive to perform and analyze (Khatamian 2011, Morgan 2004).
Other options include the use of minimally invasive interictal positron emission tomography (PET) and ictal single-photon emission computed tomography (SPECT) scans (Krakow 2008). During interictal PET, clinicians inject radioactive tracers into the patient and brain areas that do not absorb the radioactive tracers are identified as the seizure focus. Alternatively with ictal SPECT, clinicians inject radioactive tracers into the patient in order to image brain blood flow functional changes (Kim 2011) immediately after the seizure. Here, the area of the brain that absorbs the radioactive tracer is identified as the seizure focus. The timing of the injection for ictal SPECT is also very important and has to be done close to when the seizure occurred to be able to increase the sensitivity of detecting epileptogenic areas. Ictal PET and interictal SPECT both have the clinical capacity to localize seizure foci; however, despite being minimally invasive, both procedures require intravenous injection of a radio-labeled tracer into the patient and tend to reveal more regional rather than local abnormalities (Zijlmans 2007).
More invasive procedures can also be utilized for confirmation where just prior to resection, the surgeon places intradural electrodes and subdural electrode grids near the presumed seizure focus location. In order to place these intracranial electrodes, clinicians perform a craniotomy or bilateral craniotomies, which can lead to significant complications (Burneo 2006). Although these clinical methods are accurate, their complications include hemorrhages and infections, as they are invasive (Burneo 2006, Krakow 2008).
Given that the invasive techniques can have associated complications and the noninvasive and minimally invasive neuroimaging methods are sometimes inconclusive, there is a need for additional noninvasive approaches that can complement and provide further characterization. This has led to exploring resting state fMRI, a scan done while the patient is resting. This can be easily acquired (many institutions use a 5 minute acquisition protocol) along with the standard structural MRI scans in patients with different levels of disease severity and functional status. Techniques such as independent component analysis (ICA) and principle component analysis (PCA) are often used in analyzing resting state fMRI data, which allows brain network mapping that are spatiotemporally correlated due to low-frequency spontaneous oscillations of these brain regions at rest. This has led to successful mapping of language, memory, vision, and other cognitive and sensory networks at a high spatial resolution. A similar type of mapping can be performed to characterize the epileptogenic network. PCA and ICA utilize model-free analysis methods and can be used to detect interictal epileptiform discharges that are reflected in resting state fMRI BOLD activity (Morgan 2008). PCA proves to be successful only when the signal of interest is high and its frequency is slow (Sugiura 2004). ICA has been used to successfully detect fMRI activation maps that are concordant with the surgical resection (Calhoun 2003, Gu 2001, Rodionov 2007, Houdt 2015, Hunyadi 2013, Zhang 2015, Hunyadi 2015) demonstrating the power underlying a data-driven computational method. ICA generated maps involve multiple regions, strongly suggesting the presence of a network not just a seizure focus.
Temporal Clustering Analysis (TCA) is a data-driven technique that has been developed and utilized to detect fMRI signal changes in epilepsy patients that correspond to interictal epileptiform discharges and subclinical ictal discharges seen on EEG recordings (Gao 2003). In short, TCA works by first creating a histogram of the number of voxels that reach maximum intensity at each time point in the series. Then transient BOLD responses are identified that are associated with epileptiform discharges (Morgan 2009). This technique is ideal because resting fMRI is non-invasive, does not require a lot of patient interaction and can provide accurate seizure localization. When contrasted with EEG, resting fMRI is less cumbersome, has much higher spatial resolution, and is quicker to acquire (Morgan 2009, Morgan 2004).
Although Morgan, et al., (Morgan 2004) have demonstrated fairly moderate success using this method, other groups have not been able to reproduce the results reported and have attributed the TCA driven fMRI signal changes to head movement (Hamandi 2005). In addition, using simulated data Khatamian, et al., (Khatamian 2011), assessed that TCA on fMRI data worked as well as EEG-fMRI results but could only be used to either propose possible locations or validate where activity occurs as determined by other methods. Despite this limited success with TCA, we were motivated by a successful report of our group, generated by a unique patient population. Zeng et al., (Zeng 2013) conducted Regional Homogeneity (ReHo) analysis, on resting fMRI data, acquired in epilepsy patients, to successfully estimate a network responsible for seizure genesis and propagation. This unique dataset acquired from a unique patient population could prove valuable, in conducting TCA analysis, and reveal the validity of the method, in capturing epileptogenic networks, in this patient population. Surgical resection involves removal of the seizure focus and additional regions within the lobe. Rather than using TCA to identify the seizure focus, we were motivated to search for a network of regions, similar to other successful data-driven techniques.
We test the hypothesis that TCA can analyze resting fMRI data to generate useful activation maps, detailing potential epileptogenic regions of the brain that the surgeon needs to consider during presurgical planning. To that end, we utilize two-dimensional TCA (2dTCA) to analyze resting fMRI data from epilepsy patients and generate activation maps. These activation maps are then compared on an individual level to the gold standard: the known seizure focus lobe determined clinically (mri and eeg) including surrounding brain regions that were resected. Validating our hypothesis will provide a method that can complement MRI and EEG in identifying potential epileptogenic areas of the brain in epilepsy patients for surgeons to consider during resection.
Methods
Participants
Thirteen right handed epilepsy patients (6 M, age = 38.2) with varying seizure focus were selected to participate in this study (Table 1). A resting state fMRI scan was acquired for each patient between August 2010 and August 2012 at the University of Wisconsin Hospital and Clinics (UWHC), Madison. Following the standards set forth by the International League Against Epilepsy (Berg 2010), diagnosis was based on clinical EEG recordings and MRI findings. The University of Wisconsin Health Sciences Review Board approved all aspects of this study, and all participants provided voluntary written consent.
Table 1.
Overlap is presented here for all 13 patients. Overlap was determined if resection location was located within TCA activation maps
| Patient | Seizure Focus1 |
Resection Location2 |
Seizure Improved3 |
Overlap4 | MNI Coordinates5 | Corresponding Regions |
||
|---|---|---|---|---|---|---|---|---|
| x | y | z | ||||||
| 1 | left mTLE6 | left anterior temporal lobe |
y | y | −42 | −29 | +1 | left superior temporal gyrus |
| 2 | left mTLE | left amygdala hippocampus |
y | y | −38 | −41 | +2 | left middle temporal gyrus |
| 3 | left mTLE | left superior temporal gyrus |
y | y | −57 | −10 | +10 | left superior temporal gyrus |
| 4 | left mTLE | no surgery7 | N/A8 | y | −63 | −20 | +4 | left superior temporal gyrus |
| 5 | left mTLE | left anterior temporal gyrus |
y | y | −25 | +2 | −14 | left para- hippocampus |
| 6 | left mTLE | left amygdala hippocampus |
y | y | −41 | +12 | −19 | left superior temporal gyrus |
| 7 | left mTLE | left temporal lobe |
n | n | −36 | −12 | −3 | claustrum |
| 8 | right mTLE | right anterior temporal lobe |
y | y | +66 | −32 | −16 | right middle temporal lobe |
| 9 | right mTLE | right anterior temporal lobe |
y | y | +14 | +2 | +39 | right cingulate gyrus |
| 10 | left cavernous malformation9 |
left frontal craniotomy |
n | n | −1 | −26 | −9 | left red nucleus |
| 11 | right frontal encephalomalacia10 |
right anterior temporal lobe |
y | y | +29 | +11 | +49 | right middle frontal gyrus |
| 12 | left parietal encephalomalacia |
no surgery | N/A | n | N/A | N/A | N/A | N/A11 |
| 13 | right parietal | right parietal | y | y | +9 | −49 | +61 | right paracentral lobule |
Seizure Focus determined clinically using MRI and EEG
Location of the brain removed by surgeon
Clinically assessed from follow-up with neurologist or neurosurgeon 6 months and longer (up to 1 year) as no longer having seizures: y=no seizures, n=minimal to unchanged seizures
Overlap determined if Resection Location is located within TCA Activation Maps: y=matches, n=does not match at all.
MNI Coordinates (x, y, and z in mm) for most significant peak within the cluster nearest to the seizure focus
(abbrev.) mesial Temporal Lobe Epilepsy
surgery had not been performed at this time
N/A refers to no follow-up or not able to determine
Patient #10 had left hemispheric deep white matter cavernous malformation
Patient #11 had right frontal encephalomalacia secondary to trauma
TCA unable to generate activation maps
Nine of the 13 patients had mesial temporal sclerosis. All 13 patients met the following criteria during clinical evaluation:
Displayed one or more typical symptoms of mTLE and experienced complex partial seizures. In addition some patients also experienced simple partial seizures and/or secondary generalized tonic-clonic seizures.
MRI manifestations of the hippocampal sclerosis (HS), unilateral hippocampal atrophy on T1 image with associated hyperintensity on T2 fluid attenuated inverted recovery image. No structural MRI abnormality could be identified in the brain other than the hippocampal sclerosis.
Scalp EEG results showed predominantly left or right-sided interictal epileptic discharges.
Data Acquisition
During acquisition, all participants lay flat on their back with their head securely held by straps and foam pads to minimize head motion. Participants were instructed to lay as still as possible with their eyes closed, to not to think of anything specific and to not fall asleep. Five minutes eyes closed resting state images were acquired using a 3.0-Tesla scanner (GE MRI 750, Milwaukee, USA) at the UWHC, Madison.
The resting-state functional MRI data was acquired using an echo-planar imaging sequence with the following parameters: 28 axial slices, TR = 2.0 sec, TE = 30 ms, flip angle = 90°, thickness/gap = 4.0/0.0 mm, FOV = 24 × 24 cm, matrix = 64 × 64 cm, 150 volumes. A high resolution (1 mm × 1 mm × 1mm) 3D T1-weighted BRAVO anatomical MRI was acquired in an axial orientation encompassing the entire brain.
Data Analysis
Preprocessing
Using the neuroimaging software analysis, Statistical Parametric Mapping (SPM8) (Penny 2011), the functional images were preprocessed accordingly. The first three volumes were removed to account for scanner artifact. All images were then corrected for slice timing differences due to interleaved acquisition. Then realignment was used to correct for head motions and noise between acquisitions. Next the images were spatially normalized using a voxel size of [2, 2, 2] and smoothed with a FWHM Gaussian kernel of 8 mm. Participants with head motion larger than 3 mm or 3° in any of the 6 parameters (x, y, z, pitch, roll, yaw) were excluded. Each step in preprocessing is utilized in order to diminish the effects of noise on detecting changes in blood oxygenation level dependent (BOLD) responses.
2dTCA
The data series were then processed using Temporal Clustering Analysis, a data-driven technique to detect when transient changes occur in the BOLD timecourse as measured by resting fMRI. We utilized the 2dTCA toolbox, described in great detail in (Morgan 2008), to detect transient signal changes that resemble spiking activity. Briefly, each voxel, within the cortex, was categorized as either "normal" or "abnormal" by analyzing the maximum % signal change; "abnormal" voxels meet one of the following criteria:
maximum percent signal change > 0.5%, OR
maximum percent signal change > 2σ above mean of voxel's signal.
The "normal" voxels were defined to not contain BOLD signal spikes. The BOLD signal timecourse from each of the "normal" voxels were averaged to compute the global time course (GTC). The GTC was subtracted from the BOLD signal of each "abnormal" voxel to compute a signal difference. We then used a technique, called voxel cluster analysis, to map the signal difference onto a two-dimensional N×N matrix, where N was the length of the timecourse.
The diagonal of the N×N matrix was then used to identify 1–5 reference time courses (RTCs), by using the following method. If a value on the diagonal of the matrix exceeded 1.5σ above the diagonal mean, then the corresponding column of the N×N matrix as selected as a RTC. Each column identified as a RTC, consists of a group of voxels with high deviation from the GTC. With our data, 1–5 RTCs were generated for each subject, based on these criteria (Morgan 2008). To generate activation maps, we selected the RTC with the highest number of voxels, corresponding to the most widespread activation map.
The RTCs were normalized and used as regressors, along with the GTC and 6 motion parameters, in the general linear model (GLM). Using these regressors, the t-contrast for each RTC can be designated and viewed. Voxels whose time courses were similar to the regressors, had higher t-values, on the resultant activation map. The activation map displays the location of the brain that contains these abnormal spiking activities. All maps were thresholded at p < 0.05, corrected for multiple comparisons (cluster size = 5 voxels) using family-wise error (FWE) correction, provided in SPM8. Note that SPM8 displays images in "neurological" convention, which means that the right side of the image corresponds to the right side of the brain.
Results
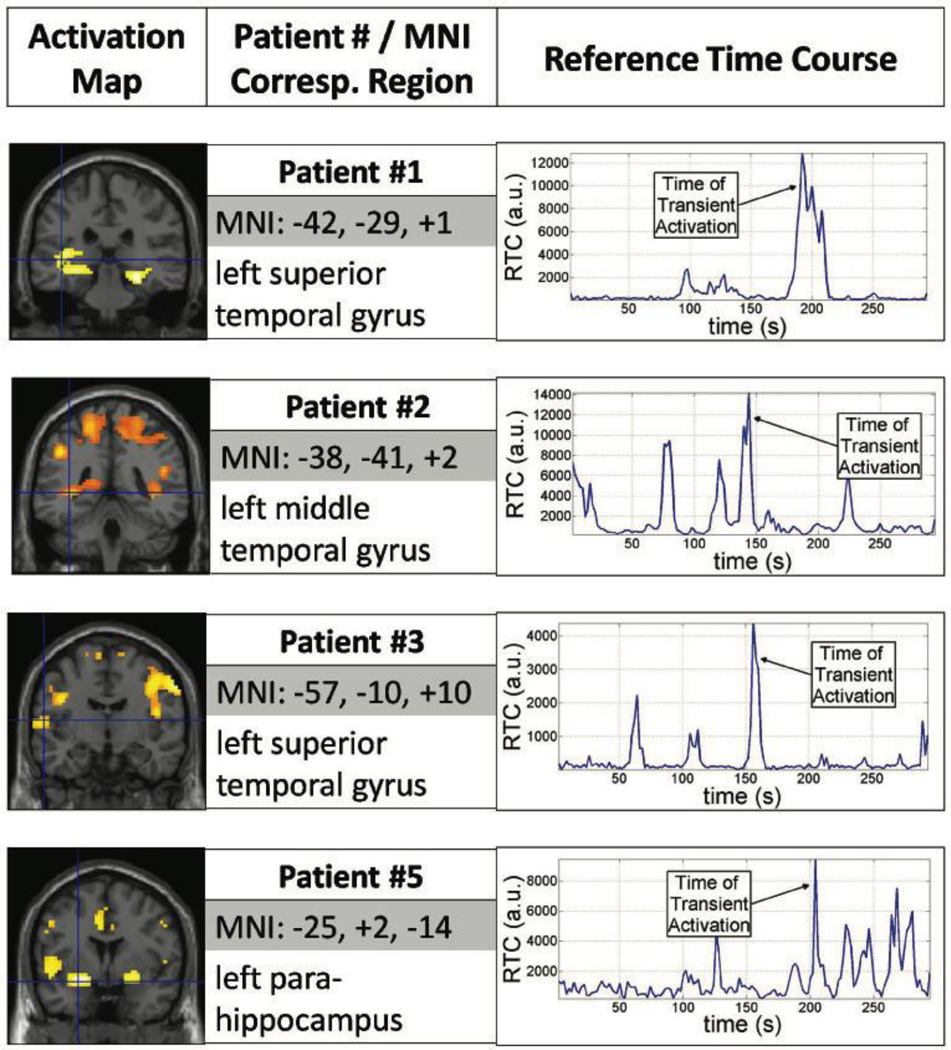
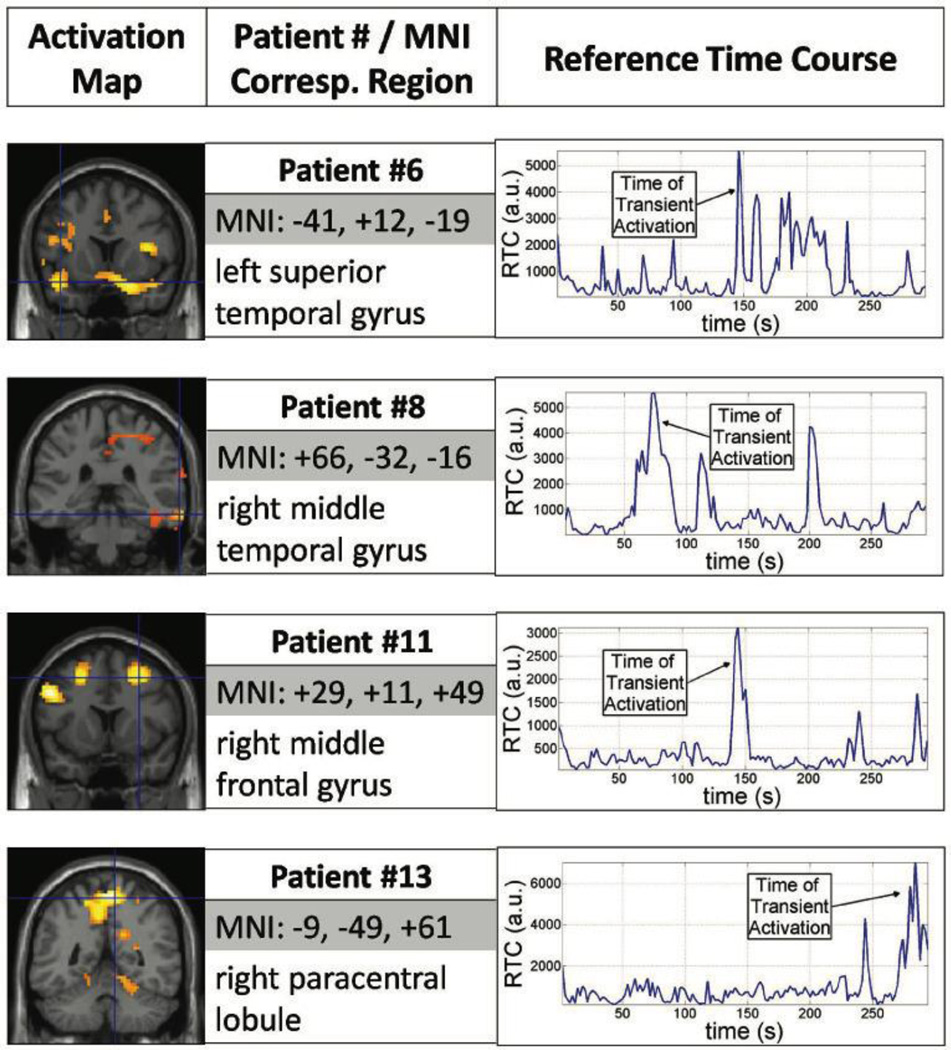
Each RTC corresponds to a different cluster of voxels that vary similarly in time; therefore, activation maps display spatial location of transient signal changes while corresponding RTCs provide a temporal measure of when the signal change is taking place. For 12 of the 13 patients analyzed, the algorithm was successful in generating RTC clusters. For patient #12, the algorithm could not converge on generating a global time course, deeming it unable to find potential epileptogenic areas. Activation maps for RTC cluster were compared to the location of the area of seizure onset determined clinically based on clinical EEG recordings and MRI findings as shown in Table 1. Results from 8 patients, shown in Figure 1 and 2, illustrate activation maps, Montreal Neurological Institute (MNI) coordinates and region name for the peak activation, and corresponding RTC used to generate activation maps.
Figure 1.
Two dimensional Temporal Clustering Analysis results for four left mTLE Patients: #1, #2, #3, and #5. Each row represents the result for each patient. In each row, activation maps (left) indicate the regions 2dTCA determined to have transient activation. The peak activation location is described (center) with MNI coordinates (in mm) and the name for the corresponding region. The reference time course used to generate the activation map is graphed (right) indicating the time transient activation occurred.
Figure 2.
Two dimensional Temporal Clustering Analysis results for another four Patients (#6, #8, #11, and #13) with varying seizure focus. Similar layout as in Figure 1: activation maps (left), peak activation coordinate (center), and reference time course (right).
In 2dTCA, the first RTC was used to identify potential epileptogenic areas in each patient. Additional RTCs correlated with other parts of the brain but were unrelated to epileptogenic regions. The activation map generated with the first RTC consisted of multiple clusters, where one of the clusters (not necessarily the biggest) was proximal to the seizure focus. In Table 1, the most significant peak within this cluster is noted, as well as eight of these patients are shown in Figure 1 and 2. Some of the clusters in the activation maps were proximal to the seizure focus but additional clusters in other parts of the brain were observed as well. Finally, the RTC (or histogram) used to generate the corresponding activation map can be used to determine retrospectively at what time during the scan, an interictal spike occurred.
In Table 1, seizure improvement assessed from follow-up with neurosurgeon or neurologist was recorded categorically as either yes or no. Yes referred to the patient not experiencing any more seizures and being seizure free, while no referred seizures still present, either some or unchanged from before surgery. In addition, concordance between resection location and corresponding TCA region was also noted categorically as either yes or no: yes regions are proximal (involving the same lobe), while no refers to regions that were distal (not involving the same lobe).
In mesial temporal seizure patients, clusters characterized by RTC activation maps were noted in the medial and superior temporal lobe, the temporal-parietal junction, the motor, and default mode network.
Discussion
Temporal Clustering Analysis on resting fMRI data has been proposed as an alternative method for localizing potential epileptogenic areas in patients with epilepsy. However, to date, only a handful of studies have investigated this new tool, with varying levels of success (Khatamian 2011, Lopes 2012, Morgan 2004). In this paper, we compared the localization of epileptogenic areas as determined by 2dTCA to that determined using standard procedures. Resting fMRI scans from 13 epilepsy patients were analyzed with 2dTCA and activation maps were generated from the first RTC. Two-dimensional TCA successfully generated activation maps for 12 of the 13 patients analyzed. Clusters proximal to the seizure focus were found in nine of these 12 patients, in addition to clusters located elsewhere in the brain. The activation map generated using 2dTCA could provide the surgeon with potential epileptogenic areas of interest. In addition to looking for abnormalities in structural MRI and abnormal spiking activity in EEG, the activation maps generated with 2dTCA could potentially highlight regions in the brain where surgeons need to pay close attention in placing subdural electrode grids for further validation.
The TCA method identified areas in varying proximity to the clinically identified seizure focus, as well as other regions that were remote but correlated with the selected RTC based on the abnormal BOLD response. These regions may all be functionally connected to the clinically identified seizure focus forming an epileptogenic network. Patients who undergo surgery often times continue to suffer from seizures, inferring that the surgeon did not remove a part of the brain where seizures are borne. These regions that appear in the activation map may be in reality part of the seizure network laying out a more complete map for the surgeon to use prior to resection, but further studies would be needed to achieve this level of confidence.
As discussed, TCA would not be able to stand alone in identifying the entire network but merely may serve as a complementary tool, laying out regions for the surgeon to investigate with other methods such as subdural electrode grids. As shown in Table 1 of the nine patients in the study that underwent resection surgery, seizure improvement and TCA and resection location concordance matched for all nine patients. One could attribute the lack of seizure improvement due to additional epileptogenic areas that were not resected. That is, given the TCA generated activation map, the surgeon could better explore regions with abnormalities by placing subdural electrode grids on the regions identified by TCA.
One limitation of the 2dTCA toolbox is that although we were able to identify a network of possible epileptogenic brain regions corresponding to the first RTC, corroboration with standard clinical assessment in terms of MRI and EEG are needed. To better understand the relationship between seizure focus and the network of regions identified by resting fMRI, studies with simultaneous recording of fMRI and EEG may be needed. Further validation could come as a result of comparing the signals recorded with subdural electrode grids from the regions highlighted in the activation maps. TCA validation will in turn improve methodology increasing the confidence necessary to assist in neurosurgery. Another limitation is the potential for neurovascular uncoupling in these patients leading to the possibility of false positive or false negative activations.
A functional connectivity analysis could help address the existence of additional regions when using 2dTCA. Reduced connectivity of mesial temporal region to frontal regions in epilepsy patients has previously been reported (Morgan 2010). Additionally, Zeng et al, used Regional Homogeneity (ReHo) in mesial temporal lobe epilepsy patients to identify network regions involved in seizure genesis and propagation (Zeng 2013). By using a more homogenous subgroup of patients with similar seizure focus, 2dTCA can first be used to determine the coordinates of the seizure focus. Then these coordinates can be used as the center-point to generate a seed region to generate functional connectivity maps showing temporal correlations with the rest of the brain. A group comparison between this homogenous epilepsy group and a group of healthy volunteers can be computed to shed light into epileptic network differences.
Based on our results, we echo the conclusions of Khatamian, et al., (Khatamian 2011) that 2dTCA has the potential to add complementary information regarding the localization of seizure focus and the epileptogenic network in each individual subject. For example, when EEG and structural MRI are in disagreement, TCA could be used to help assess potential epileptogenic areas. Likewise, when no apparent lesions are observable in an MRI scan but EEG is showing abnormal activity over a part of the brain, TCA can be quickly implemented to help determine the location. Further, resting state fMRI scans can be collected easily and quickly in 5 minute scans, possibly following the structural MRI scan which is routinely collected in epilepsy patients.
Conclusion
Analyzing resting fMRI data using novel data-driven techniques, like TCA, has the potential to be utilized concurrently with standard clinical methods in order to localize an epileptic network. The data for thirteen patients was analyzed using TCA with 12 of 13 patients yielding a network of potential epileptogenic areas. The location of the nearest cluster from these activation maps varied in proximity to the area of seizure onset determined clinically. The approach adopted here has the potential to be used clinically as it is non-invasive like EEG, computationally quick to implement, and can be used in patients with different levels of disease severity, and functional status.
Acknowledgments
Financial Support
The project described is supported by NLM 5T15LM007359, NIH NIMH RC1MH090912-01 Challenge Grant, American Heart Association Grants, NCATS grant 9U54TR000021, NIGMS R25GM083252, 5RC1MH090912, NIH ICTR KL2 Scholar Mentored Career Development Award, UW ICTR NIH/UL1RR025011 Pilot Grant, American Society of Neuroradiology NERF Scholar Award, Coulter Foundation Grant, UW Milwaukee-Madison Intercampus Grant, Shapiro Grants, UW NTP, CNTP training grants, and UW Startup Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010;51(4):676–685. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- Burneo JG, Steven DA, McLachlan RS, Parrent AG. Morbidity associated with the use of intracranial electrodes for epilepsy surgery. Can J Neurol Sci. 2006;33(2):223–227. doi: 10.1017/s0317167100005023. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Hansen LK, Larsen J, Pekar JJ. Ica of functional mri data: an overview. 2003 [Google Scholar]
- Gao JH, Yee SH. Iterative temporal clustering analysis for the detection of multiple response peaks in fMRI. Magn Reson Imaging. 2003;21(1):51–53. doi: 10.1016/s0730-725x(02)00627-6. [DOI] [PubMed] [Google Scholar]
- Gu H, Engelien W, Feng H, Silbersweig DA, Stern E, Yang Y. Mapping transient, randomly occurring neuropsychological events using independent component analysis. NeuroImage. 2001;14(6):1432–1443. doi: 10.1006/nimg.2001.0914. [DOI] [PubMed] [Google Scholar]
- Hamandi K, Salek Haddadi A, Liston A, Laufs H, Fish DR, Lemieux L. fMRI temporal clustering analysis in patients with frequent interictal epileptiform discharges: comparison with EEG-driven analysis. Neuroimage. 2005;26(1):309–316. doi: 10.1016/j.neuroimage.2005.01.052. [DOI] [PubMed] [Google Scholar]
- Hunyadi B, Tousseyn S, Dupont P, Van Huffel S, De Vos M, Van Paesschen W. A prospective fmri-based technique for localising the epileptogenic zone in presurgical evaluation of epilepsy. NeuroImage. 2015;113:329–339. doi: 10.1016/j.neuroimage.2015.03.011. [DOI] [PubMed] [Google Scholar]
- Hunyadi B, Tousseyn S, Mijović B, Dupont P, Van Huffel S, Van Paesschen W, De Vos M. Ica extracts epileptic sources from fmri in eeg-negative patients: a retrospective validation study. PloS one. 2013;8(11):e78796. doi: 10.1371/journal.pone.0078796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatamian YB, Fahoum F, Gotman J. Limits of 2D-TCA in detecting BOLD responses to epileptic activity. Epilepsy Res. 2011 doi: 10.1016/j.eplepsyres.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Mountz JM. SPECT Imaging of Epilepsy: An Overview and Comparison with F-18 FDG PET. Int J Mol Imaging. 2011;2011:813028. doi: 10.1155/2011/813028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi E, Bagshaw AP, Benar CG, Aghakhani Y, Andermann F, Dubeau F, et al. Temporal and extratemporal BOLD responses to temporal lobe interictal spikes. Epilepsia. 2006;47(2):343–354. doi: 10.1111/j.1528-1167.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- Krakow K. Imaging epileptic activity using functional MRI. Neurodegener Dis. 2008;5(5):286–295. doi: 10.1159/000135613. [DOI] [PubMed] [Google Scholar]
- Lopes R, Lina JM, Fahoum F, Gotman J. Detection of epileptic activity in fMRI without recording the EEG. Neuroimage. 2012;60(3):1867–1879. doi: 10.1016/j.neuroimage.2011.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan VL, Price RR, Arain A, Modur P, Abou-Khalil B. Resting functional MRI with temporal clustering analysis for localization of epileptic activity without EEG. Neuroimage. 2004;21(1):473–481. doi: 10.1016/j.neuroimage.2003.08.031. [DOI] [PubMed] [Google Scholar]
- Morgan VL, Li Y, Abou-Khalil B, Gore JC. Development of 2dTCA for the detection of irregular, transient BOLD activity. Hum Brain Mapp. 2008;29(1):57–69. doi: 10.1002/hbm.20362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan VL, Gore JC. Detection of irregular, transient fMRI activity in normal controls using 2dTCA: comparison to event-related analysis using known timing. Hum Brain Mapp. 2009;30(10):3393–3405. doi: 10.1002/hbm.20760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan VL, Gore JC, Abou-Khalil B. Functional epileptic network in left mesial temporal lobe epilepsy detected using resting fMRI. Epilepsy Res. 2010;88(2–3):168–178. doi: 10.1016/j.eplepsyres.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny WD, Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE. Statistical parametric mapping: the analysis of functional brain images: the analysis of functional brain images. Academic press; 2011. [Google Scholar]
- Rodionov R, De Martino F, Laufs H, Carmichael D, Formisano E, Walker M, Lemieux L. Independent component analysis of interictal fmri in focal epilepsy: comparison with general linear model- based eeg-correlated fmri. Neuroimage. 2007;38(3):488–500. doi: 10.1016/j.neuroimage.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Salek-Haddadi A, Merschhemke M, Lemieux L, Fish DR. Simultaneous EEG-Correlated Ictal fMRI. Neuroimage. 2002;16(1):32–40. doi: 10.1006/nimg.2002.1073. [DOI] [PubMed] [Google Scholar]
- Sugiura M, Watanabe J, Maeda Y, Matsue Y, Fukuda H, Kawashima R. Different roles of the frontal and parietal regions in memory-guided saccade: a PCA approach on time course of BOLD signal changes. Hum Brain Mapp. 2004;23(3):129–139. doi: 10.1002/hbm.20049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houdt PJ, Ossenblok PP, Colon AJ, Hermans KH, Verdaasdonk RM, Boon PA, de Munck JC. Are epilepsy-related fmri components dependent on the presence of interictal epileptic discharges in scalp eeg? Brain topography. 2015;28(4):606–618. doi: 10.1007/s10548-014-0407-1. [DOI] [PubMed] [Google Scholar]
- Zeng H, Pizarro R, Nair VA, La C, Prabhakaran V. Alterations in regional homogeneity of resting-state brain activity in mesial temporal lobe epilepsy. Epilepsia. 2013 doi: 10.1111/epi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CH, Lu Y, Brinkmann B, Welker K, Worrell G, He B. Lateralization and localization of epilepsy related hemodynamic foci using presurgical fmri. Clinical Neurophysiology. 2015;126(1):27–38. doi: 10.1016/j.clinph.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans M, Huiskamp G, Hersevoort M, Seppenwoolde JH, van Huffelen AC, Leijten FS. EEG-fMRI in the preoperative work-up for epilepsy surgery. Brain. 2007;130(Pt 9):2343–2353. doi: 10.1093/brain/awm141. [DOI] [PubMed] [Google Scholar]