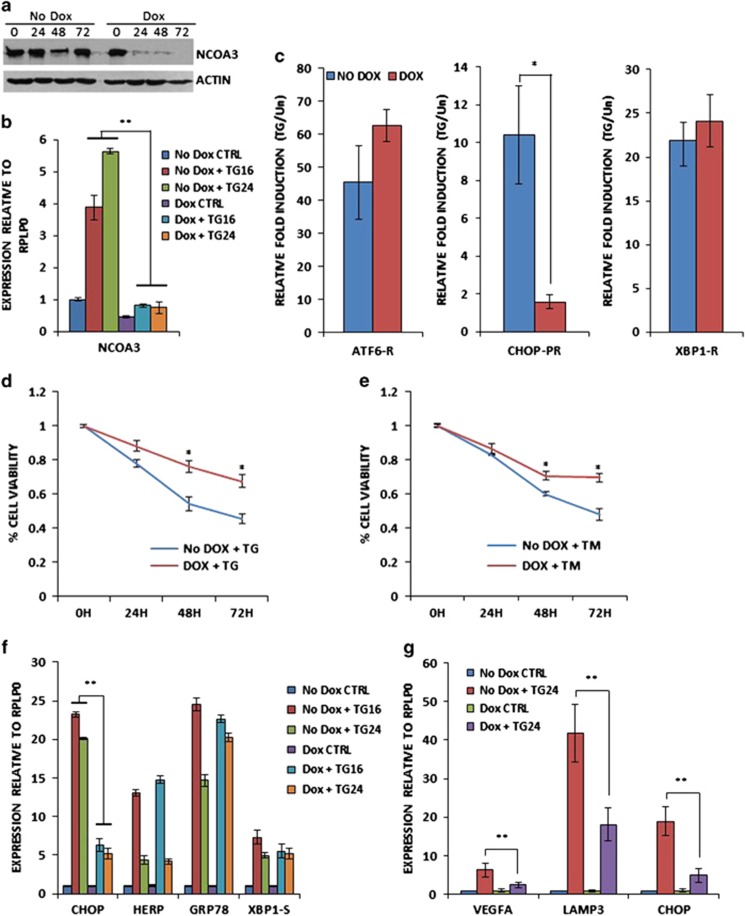
Figure 4.
NCOA3 is required for EnR stress-induced activation of the PERK–ATF4–CHOP axis. (a) pTRIPZshNCOA3-MCF7 cells were either untreated (No Dox) or treated (Dox) with (500 ng/ml) of doxycycline for indicated time points. Upper panel, equivalent amounts of cell lysates were resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and immunoblotting was performed using antibodies against NCOA3 and β-actin. (b) pTRIPZshNCOA3-MCF7 cells were either untreated (CTRL) or treated with (1.0 μm) TG for indicated time points in the absence and presence of doxycycline. The expression level of NCOA3 was quantified by real-time RT–PCR, normalizing against RPLP0. Error bars represent mean±s.d. from three independent experiments performed in triplicate. (c) pTRIPZshNCOA3-MCF7 cells were transfected with the indicated UPR pathway reporter genes (ATF6-R, CHOP-PR, XBP1-R). Transfected cells were treated with TG (1.0 μm) in the absence and presence of doxycycline for 24 h. Normalized luciferase activity (Firefly/Renilla) relative to untreated control is shown. Error bars represent mean±s.d. from three independent experiments performed in duplicate. (d, e) pTRIPZshNCOA3-MCF7 cells were untreated (CTRL) or treated with (1.0 μm) TG and (1.0 μg/ml) TM in the absence and presence of (500 ng/ml) of doxycycline for indicated time points. Line graphs show the absorbance in cells at the indicated time points after the treatment. Error bars represent mean±s.d. from three independent experiments performed in triplicate. (f, g) pTRIPZshNCOA3-MCF7 cells were either untreated (CTRL) or treated with (1.0 μm) TG for indicated time points in the absence and presence of doxycycline. The expression level of CHOP, HERP, GRP78, XBP1-S, VEGFA and LAMP3 was quantified by real-time RT–PCR, normalizing against RPLP0. Error bars represent mean±s.d. from three independent experiments performed in triplicate. *P<0.05, two-tailed unpaired t-test comparing TG-induced samples; **P<0.05, two-tailed unpaired t-test comparing respective time points.