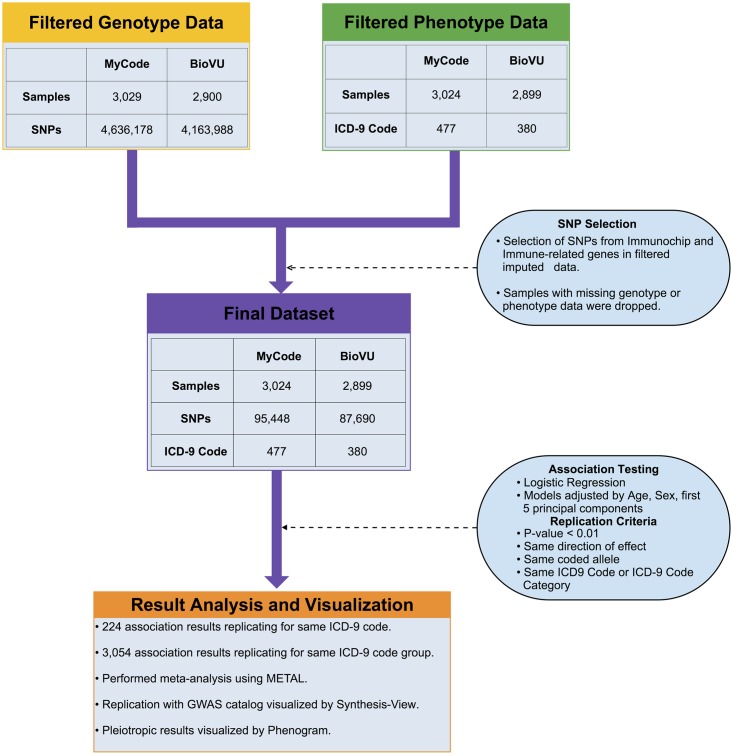
Fig 1. Overview of PheWAS with Immune Variants.
This flow chart provides an overview of the steps taken to perform PheWAS between immune variants and ICD-9 diagnosis codes. The final testing dataset (purple) was formed by selecting SNPs from our array data that also exist on Immunochip and/or are within immune-related genes (yellow) and removing samples with missing genotypic or phenotypic data (green). Comprehensive associations were calculated between all final dataset SNPs and ICD-9 code based case/control status using logistic regression, with all models adjusted for age, sex and first five principal components. Replication was sought following both an exact ICD-9 code and a category ICD-9 code approach following the specified criteria. Pooled analysis was performed for both approaches using METAL. See S1 Fig for the full workflow from imputation through quality control, association testing, and replication for this study.