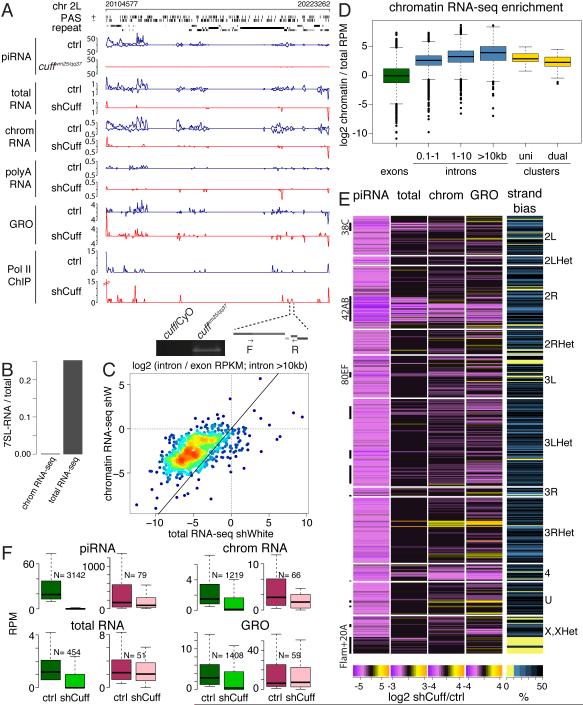
Fig. 1. Cuff deficiency eliminates the majority of transcripts from piRNA clusters.
(A) Cuff depletion leads to diminished piRNA cluster transcription. Shown are profiles of piRNA, total cell and poly(A)-selected RNA-seq, chromatin RNA-seq, global nuclear run-on seq (GRO-seq), and RNA polymerase II ChIP-seq from ovaries of control and cuff depleted flies on the dual-strand piRNA cluster 38C. The distribution of predicted poly(A) signals on plus and minus genomic strands is shown in the poly(A) signal (PAS) tracks. Below is an expansion of the indicated region from which poly-adenylated transcripts detected by RT-PCR using oligo-dT primer are produced in cuffwm25/qq37 flies.
(B) Chromatin-associated RNA sequencing libraries are depleted of signal recognition particle (7SL) RNA.
(C) Chromatin-associated RNAs are enriched in intronic sequences as measured by the ratio of intronic to exonic RPKM signals. Solid line indicates no difference between standard and chromatin RNA-seq.
(D) piRNA cluster transcripts are enriched in chromatin-associated RNA.
(E) Cuff suppresses primary transcripts from Cuff-depended piRNA producing loci. Heatmap shows signal change in Cuff-depleted versus control ovaries for small RNA, total RNA, chromatin-associated RNA and GRO-seq in genomic windows that produce piRNA in a Cuff-dependent manner. Intervals overlapping uni-strand piRNA clusters that are not affected by Cuff depletion are shown for comparison (Flam+20A).
(F) Box plots show the distribution of piRNA, chromatin-associated RNA, total RNA and GRO signals in control and Cuff-depleted ovaries. Green boxes corresponds to all genomic regions that produce piRNA in Cuff-dependent manner; pink boxes correspond to windows overlapping uni-strand piRNA clusters (flamenco and 20A). See also Figure S1.