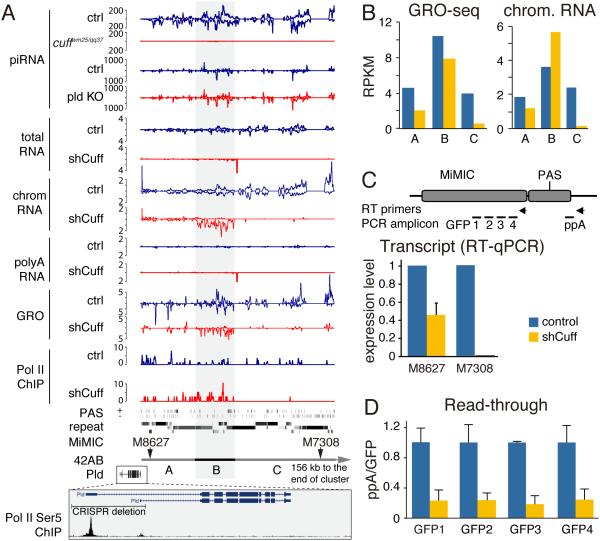
Fig. 2. Cuff suppresses transcription termination at canonical poly(A) sites.
(A) Profiles of different libraries over the 42AB cluster which is further divided into thee regions, A, B and C. Below shown are the locations of two insertions of artificial MiMIC transposons, and the site of CRISPR-mediated deletion of promoters of the Pld gene, which flanks the 42AB cluster. The entire cluster is shown on Figure S2A.
(B) Cuff depletion leads to reduced transcription of the 42AB cluster. Normalized densities of GRO-seq and chromatin RNA-seq signals are calculated for three regions shown in (A).
(C) Depletion of Cuff has a different effect on two promoterless MiMIC insertions located at different positions in the 42AB cluster. Shown is the amount of MiMIC RNA in control and cuff-depleted (shCuff) ovaries as determined by RT-qPCR. Error bars show standard errors from three biological replicates.
(D) Depletion of Cuff suppresses read-through transcription downstream of the canonical poly(A) site of the M8627 insertion. Amounts of RNA upstream (GFP1-4) and downstream (ppA) of the poly(A) site were quantified by strand-specific RT-qPCR and the ratios of read-through (ppA/GFP1-4) transcripts were plotted. RT primers and qPCR amplicons are shown in panel C. Error bars show standard errors from three biological replicates. See also Figure S2B.