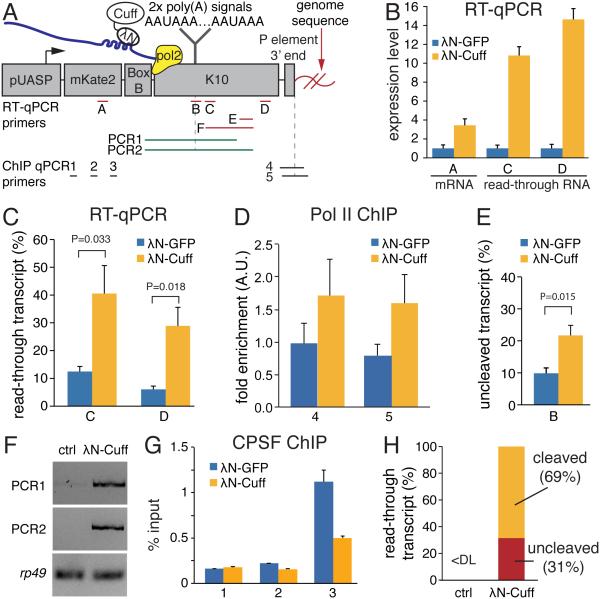
Fig. 4. Tethering of Cuff to a heterologous mRNA reporter suppresses termination and poly(A) site cleavage.
(A) Schematic diagram of the reporter used to study the effect of Cuff recruitment to RNA. Cuff fused to the λN peptide, which binds BoxB hairpins in reporter RNA, was co-expressed with the reporter in ovaries.
(B) Cuff tethering leads to increased reporter transcript levels. λN-Cuff or control λN-GFP were co-expressed with the reporter in fly ovaries and RNA levels from three regions shown on (A) were quantified by RT-qPCR. Error bars show standard errors of four biological replicates.
(C) Tethering of Cuff increases read-through transcription of the reporter pre-mRNA as measured by the fraction of read-through transcripts relative to total reporter RNA. Error bars are standard errors from four biological replicates. P values were calculated by t-test. See also Figure S4.
(D) Tethering of Cuff suppresses Pol II termination as measured by Pol II ChIP-qPCR downstream of the poly(A) signals (fragments 4 and 5 on panel A). Error bars show standard errors of two biological replicates.
(E) Tethering of Cuff inhibits cleavage at poly(A) sites. Shown is the fraction of unprocessed transcripts calculated as described on Figure S5. Error bars are standard errors from four biological replicates. P was calculated by t-test.
(F) Unprocessed reporter transcripts that span the poly(A) site are detected by RT-PCR upon λN-Cuff tethering (amplified regions are shown in panel A).
(G) Tethering of Cuff suppresses binding of CPSF complex as measured by Cpsf73 ChIP-qPCR on different portions of the reporter shown on (A). Error bars show standard errors of four biological replicates.
(H) The majority of read-through transcripts formed upon Cuff tethering are cleaved near poly(A) sites. The fractions of cleaved and unprocessed RNA in two biological replicas were determined using standard curves shown on Figure S4 and S5.