Abstract
Inorganic phosphate release, [Pi], is often measured in an enzymatic reaction in a high throughput setting. Based on the published mechanism, we designed a protocol for our screening for inhibitors of SAICAR synthetase (PurC), and we found a gradual increase in [Pi] in positive control samples over the course of the day. Further investigation indicated that hydrolysis of ATP catalyzed by PurC, rather than substrate-related phosphate release, was responsible for a partial contribution to the signals in the control samples. Thus substrate-independent ATPase activity may complicate high throughput screening.
Keywords: Malachite Green, high throughput screening, substrate-independent ATPase activity
SAICAR synthetase (PurC) is involved in de novo purine biosynthesis, and is present in most forms of life. As an individual enzyme in many bacterial species, PurC converts L-Asp, 4-carboxy-5-aminoimidazole ribonucleotide (CAIR) and ATP to 4-(N-succino)-5-aminoimidazole-4-carboxamide ribonucleotide (SAICAR), ADP and Pi [1–3]. In human, a bi-functional enzyme, PAICS, harbors a PurC domain [4]. PurC has been suggested as a viable target for antimicrobials [5] as well as for chemo-therapeutics [6]. We implemented a Malachite Green assay (MGA) in a high throughput screening (HTS) setup to identify compounds that inhibit the activity of Bacillus anthracis PurC (BaPurC). MGA has long been used to monitor the activities of enzymes that release inorganic phosphate by detecting the phosphomolybdate-Malachite Green complex [7,8], and is a common method for the primary screening of inhibitors against enzymes with NTPase activity [9–11].
Our assay was based on the published mechanism, suggesting direct activation of the carboxylate of CAIR by the γ-phosphate of ATP, leading to the formation of products, SAICAR, ADP and Pi [2,15]. After taking into consideration the Km and Kcat values [15], the assay was optimized for HTS and performed in clear flat bottom 384 well plates in 50 mM Tris buffer at pH 8.0 with 100 mM NaCl, 1 mM MgCl2, 0.01% Triton, and 0.1 mg/mL BSA (Assay Buffer). Each sample well contained purified BaPurC [12] (0.5 µM), L-Asp (2.4 mM), ATP (40 µM) and a specific compound (29 µM) from a small-molecule library (ChemBridge). The compounds were in DMSO, and the DMSO final concentration in the sample was 0.3%. About 40 plates were prepared and screened per day. For each plate, just prior signal detection, the reactions were initiated with the addition of the critical substrate CAIR (20 µM) and allowed to react for 6 min at 25 °C, followed by the addition and 10 min incubati on of Malachite Green dye. The Malachite Green solution was either prepared according to published methods [13,14], or purchased (BioAssay Systems; POMG-25H). The absorbance values at 633 nm of samples in each well were read with a plate reader (Tecan Infinite 200 Pro microplate reader; University of Illinois at Chicago RRC HTS facility), and converted to [Pi] with a calibration curve. We also included positive controls in 32 wells that contained no compound, and negative controls in 32 wells that contained no BaPurC. The overall Z'-factor of the completed runs was 0.85, validating the quality of the assay [16].
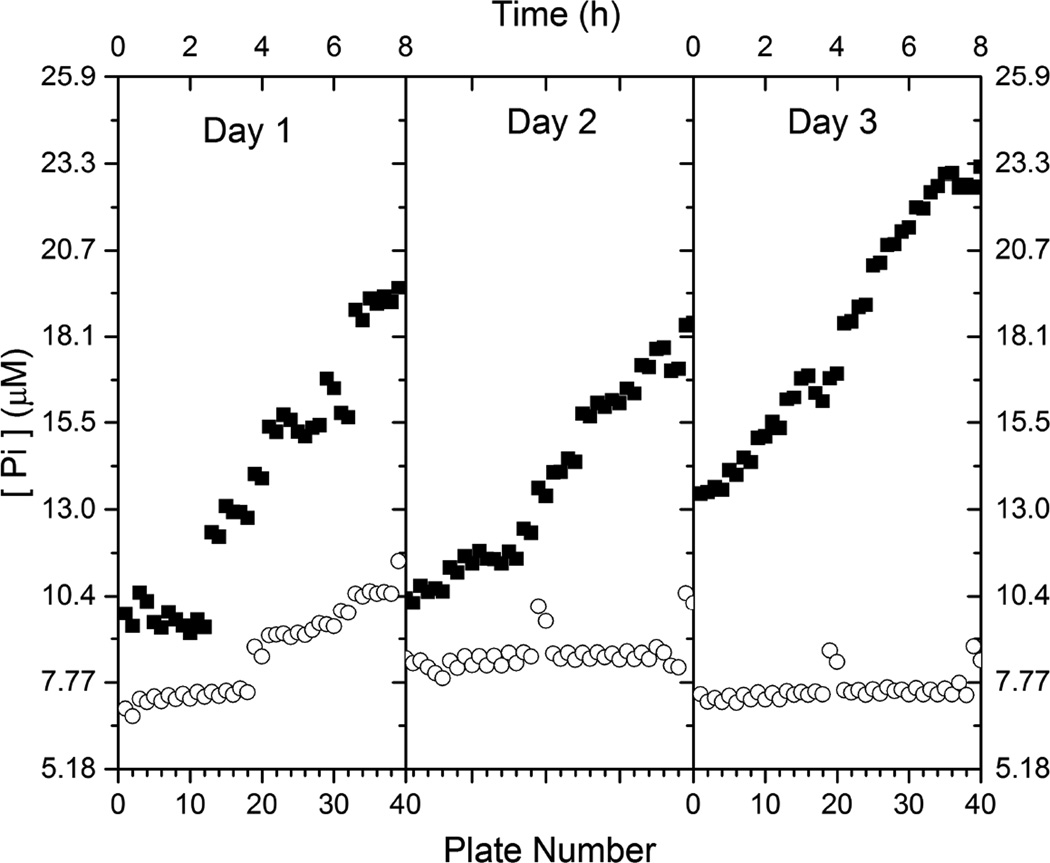
In our data analysis, an unexpected trend was noticed. The signals of the positive controls (BaPurC and substrates) did not stay constant during the 8 hr time period each day; instead they steadily increased (Fig 1). The signals of the negative controls (no BaPurC) stayed relatively constant. The reaction scheme is shown in Fig 2 (inset). The results suggested that the signals (Pi concentrations) were not from non-enzymatic ATP hydrolysis, but from a slow enzyme-catalyzed ATP hydrolysis. Intrinsic ATPase activity has been well characterized in protein kinases and is not a new phenomenon [17–20], but would be the first study of this activity in a PurC enzyme.
Figure 1. [Pi] of positive controls of BaPurC MGA HTS show a general increase as a function of time.
The [Pi] values, from Malachite Green absorbance values, of control samples (n = 32) in each well of the HTS plate, detecting the enzymatic activity of BaPurC. A total of 40 plates were run in a time period of 8 h. Each plate consisted of 32 wells for control samples. The positive control samples (■) consisted of BaPurC (0.5 µM), ATP (40 µM) and L-Asp (2.4 mM) in 50 mM Tris buffer at pH 8.0 with 100 mM NaCl, 1 mM MgCl2, 0.01% Triton, 0.3% DMSO and 0.1 mg/mL BSA. Shortly before each plate was ready for signal detection, CAIR (20 µM) was added and incubated for 6 min. Malachite Green solution was then added and incubated for 10 min. The data for 3 different days are shown. All data of the positive control samples show a gradual rise in the 8 h time period for each of the 3 days. However, the negative control samples (○), which did not include BaPurC, exhibited little or no increase in the [Pi] values.
Figure 2. Incubation time and order of assay component addition reveal substrate independent ATP hydrolysis in BaPurC.
Tris buffer (50 mM) at pH 8.0 with 100 mM NaCl and 1 mM MgCl2 was used as Assay Buffer for all samples. All three samples in Set A (■) consisted of BaPurC (0.5 µM), ATP (40 µM) and L-Asp (2.4 mM). After a sample was incubated to 1, 4 or 8 h at 25 °C, CAIR (20 µM) was added and incubated for 6 min, followed by Malachite Green addition and incubation for 10 min. Samples in Set B (□), CAIR was replaced with buffer, i.e. no CAIR in the samples. Samples in Set C (●) consisted of CAIR (20 µM), ATP (40 µM) and L-Asp (2.4 mM). After a sample was incubated to 1, 4 or 8 h at 25 °C, BaPurC (0.5 µM) was added and incubated for 6 min, followed by Malachite Green addition and incubation for 10 min. For samples in Set D (○), BaPurC was replaced with buffer (i.e. no BaPurC in samples; similar to the negative control in HTS). The Pi concentration in each sample was obtained from a calibration curve using the absorbance values at 630 nm. For all samples, the data were the averaged values of n = 5 – 10.
To further investigate this possibility, samples and controls mimicking HTS conditions were prepared. Set A samples in Assay Buffer consisted of BaPurC (0.5 µM), ATP (40 µM) and L-Asp (2.4 mM). After the samples were incubated to 1, 4 or 8 h at 25 °C, CAIR (20 µM) was added and incubated for 6 min, followed by Malachite Green addition and incubation for 10 min. Set B samples were similar to Set A samples except that CAIR was replaced with buffer, i.e. no CAIR in samples. Set C samples consisted of CAIR (20 µM), ATP (40 µM) and L-Asp (2.4 mM). After the samples were incubated to 1, 4 or 8 h at 25 °C, BaPurC (0.5 µM) was added and incubated for 6 min, followed by Malachite Green addition and incubation for 10 min. Set D samples were similar to Set C samples except BaPurC was replaced with buffer, i.e. no BaPurC in samples. The absorbance values at 630 nm were obtained with a plate-reader (Perkin-Elmer Victor3 V 1420 multilabel counter), and converted to the Pi concentrations with a calibration curve.
Results from samples in Set A showed an increase in [Pi] as a function of time, from ~7 µM at t = 1 hr, to ~19 µM at t = 8 hr (Fig 2), similar to those observed in HTS. In the absence of CAIR (samples in Set B), [Pi] increased from ~3 µM at t = 1 hr, to ~15 µM at t = 8 hr. When BaPurC was added just shortly before measurement (samples Set C), a relatively constant [Pi], at ~7 µM, was observed. For samples without the enzyme (Set D), a relatively constant [Pi], at ~ 3 µM, was observed. Since 40 µM ATP was present in the reaction samples, we expect that the signals in samples in Sets A and B will continue to increase beyond 8 h, until all ATP is converted.
Our findings also support our recent proposed mechanism showing that PurC indeed catalyzes ATP hydrolysis to allow a phosphate relay between ATP and conserved Glu residues to activate CAIR [3].
In HTS measuring enzyme activities that involve multiple substrates and co-factors, the decisions on when to introduce which substrates and co-factors to the reaction mixture may be complicated. A logical approach is to design protocols based on published mechanisms. However, as shown in this study, these protocols still need to be carefully tested to provide data that can be meaningfully analyzed. As indicated in recent publications, HTS data sets often have inherent systematic and random error, which may lead to false-positive or false-negative results, and the diversity of the features of different enzymes prevents unification of assay conditions [21–23]. Among many protocols, the assay mixture and blank preparation and the choice of the assay time are crucial to avoid frequent and trivial but costly errors [23].
In this report, we point out potential problems in detecting [Pi] in systems where ATP was a co-factor and not a substrate. Control samples should include samples detecting potential enzyme induced ATP hydrolysis. To our knowledge this is the first detailed analysis showing the pitfall of substrate-independent ATPase activity in PurC systems, and we caution others to take similar enzymatic event into consideration in future experimentation.
Acknowledgments
We would like to thank Shahila Mehboob and Michael Johnson at the UIC Center of Pharmaceutical Biotechnology for the BaPurC plasmids. The work was supported, in part, by a grant from the National Institutes of Health (U19 AI-56575).
Abbreviations
- CAIR
4-carboxy-5-aminoimidazole ribonucleotide
- HTS
high throughput screening
- MGA
malachite green assay
- PurC
succinoaminoimidazolecarboxamide ribonucleotide synthethase
- SAICAR
succinoaminoimidazolecarboxamide ribonucleotide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levdikov VM, Barynin VV, Grebenko AI, Melik-Adamyan WR, Lamzin VS, Wilson KS. The structure of SAICAR synthase: an enzyme in the de novo pathway of purine nucleotide biosynthesis. Structure. 1998;6:363–376. doi: 10.1016/s0969-2126(98)00038-0. [DOI] [PubMed] [Google Scholar]
- 2.Ginder ND, Binkowski DJ, Fromm HJ, Honzatko RB. Nucleotide complexes of Escherichia coli phosphoribosylaminoimidazole succinocarboxamide synthetase. J. Biol. Chem. 2006;281:20680–20688. doi: 10.1074/jbc.M602109200. [DOI] [PubMed] [Google Scholar]
- 3.Wolf NM, Abad-Zapatero C, Johnson ME, Fung LW-M. Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg2+, AIR and Asp. Acta Cryst. 2014;D70:841–850. doi: 10.1107/S139900471303366X. [DOI] [PubMed] [Google Scholar]
- 4.Li S-X, Tong Y-P, Xie X-C, Wang Q-H, Zhou H-N, Zhang Z-Y, Gao W, Li S-G, Zhang XC, Bi R-C. Octameric structure of the human bifunctional enzyme PAICS in purine biosynthesis. J. Mole. Bio. 2007;366:1603–1614. doi: 10.1016/j.jmb.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 5.Yuan Z, Wang L, Sun S, Wu T, Qian W. Genetic and proteomic analysis of a Xanthomonas campestris pv. campestris purC mutant deficient in purine biosynthesis and virulence. J. Genet. Genom. 2013;40:473–487. doi: 10.1016/j.jgg.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Keller KE, Tan IS, Lee Y-S. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itaya K, Ui M. A new micromethod for the colorimetric determination of inorganic phosphate. Clinica Chimica Acta. 1966;14:361–366. doi: 10.1016/0009-8981(66)90114-8. [DOI] [PubMed] [Google Scholar]
- 8.Kiani FA, Fisher S. In: Molecular catalysts: structure and functional design. Gade LH, Hofmann P, editors. Weinheim, Germany: Wiley-Vch; 2014. Chap.17. [Google Scholar]
- 9.Bharat A, Blanchard JE, Brown ED. A high-throughput screen of the GTPase activity of Escherichia coli EngA to find an inhibitor of bacterial ribosome biogenesis. J. Biomol. Screen. 2013;18:830–836. doi: 10.1177/1087057113486001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang L, Bertelsen EB, Wisén S, Larsen EM, Zuiderweg ERP, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of molecular chaperone DnaK. Anal. Biochem. 2008;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 11.Rowlands MG, Newbatt YM, Prodromou C, Pearl LH, Workman P, Aherne W. High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Anal. Biochem. 2004;327:176–183. doi: 10.1016/j.ab.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 12.Tuntland ML, Wolf NM, Fung LW-M. Differences in the purification and solution properties of PurC gene products from Streptococcus pneumoniae and Bacillus anthracis. Protein Expr. Purif. 2015;114:143–148. doi: 10.1016/j.pep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Pegan SD, Tian Y, Sershon V, Mesecar AD. A universal, fully automated high throughput screening assay for pyrophosphate and phosphate release from enzymatic reactions. Comb. Chem. & High Throughput Screen. 2010;13:27–38. doi: 10.2174/138620710790218203. [DOI] [PubMed] [Google Scholar]
- 14.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 15.Nelson SW, Binkowski DJ, Honzatko RB, Fromm HJ. Mechanism of action of Escherichia coli phosphoribosylaminoimidazolesuccinocarboxamide synthetase. Biochem. 2005;44:766–774. doi: 10.1021/bi048191w. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J-H, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 17.Kaji A, Colowick SP. Adenosine triphosphatase activity of yeast hexokinase and its relation to the mechanism of the hexokinase reaction. J. Biol. Chem. 1965;240:4454–4462. [PubMed] [Google Scholar]
- 18.Paudel HK, Carlson GM. The ATPase activity of phosphorylase kinase is regulated in parallel with its protein kinase activity. J. Biol. Chem. 1991;266:16524–16529. [PubMed] [Google Scholar]
- 19.Hurley JH. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu. Rev. Biophys. Biomol. Struct. 1996:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 20.Kashem MA, Nelson RM, Yingling JD, Pullen SS, Prokopowicz AS, III, Jones JW, Wolak JP, Rogers GR, Morelock MM, Snow RJ, Homon CA, Jakes S. Three mechanistically distinct kinase assays compared: measurement of intrinsic ATPase activity identified the most comprehensive set of ITK inhibitors. J. Biomol. Screen. 2007;12:70–83. doi: 10.1177/1087057106296047. [DOI] [PubMed] [Google Scholar]
- 21.Mangat CS, Bharat A, Gehrke SS, Brown ED. Rank ordering plate data facilitates data visualization and normalization in high-throughput screening. J. Biomol. Screen. 2014;19:1314–1320. doi: 10.1177/1087057114534298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acker MG, Auld DS. Considerations for the design and reporting of enzyme assays in high-throughput screening applications. Persp. in Sci. 2014;1:56–73. [Google Scholar]
- 23.Bisswanger H. Enzyme assays. Persp. in Sci. 2014;1:41–55. [Google Scholar]