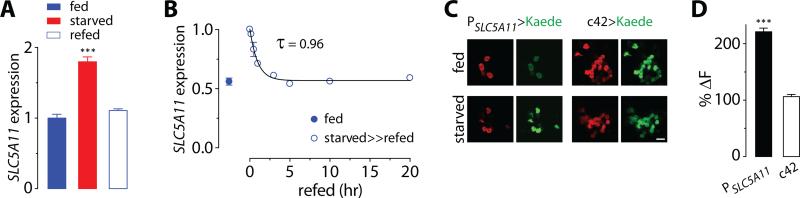
Fig. 3. Starvation increases the expression of SLC5A11 transcript.
(A) Quantitative real-time PCR assay for SLC5A11 transcripts using the SYBR green method from the brains of flies that were food deprived for 5 hours (fed) and for 22 hours (starved), and had been deprived for 22 hours, but were refed for 24 hours (refed). The transcript levels in starved and refed flies are normalized to those in fed flies. GAPDH is used as a reference (n = 9, three independent biological replicates; ***P < 0.001 vs. fed). (B) SLC5A11 transcript levels of fed flies (fed), and 22-hr starved flies that were refed for 0, 10, 30, 60, 180, 300, 600 and 1,200 minutes (starved>>refed). The transcript levels of the starved>>refed flies are normalized to those in starved flies. The rate constant is predicted by a single exponential decay function. (C) Starvation-induced transcriptional activity of the SLC5A11 promoter in SLC5A11 neurons is visualized by monitoring de novo synthesis of green fluorescence in flies carrying PSLC5A11-GAL4 and UAS-Kaede. After preexisting green fluorescence was photoconverted to red fluorescence, the flies were reared in either fed or starved conditions prior to fluorescence imaging. Scale bar, 20μm. (D) Relative amount of newly synthesized green KAEDE in SLC5A11 neurons in starved condition. The values were calculated by normalizing de novo KAEDE to preexisting red KAEDE (ΔF) followed by normalizing the measurements from starved condition to those from fed condition (%ΔF; n = 26-42; ***P < 0.001). Note that the control c42-Gal4 labeled neurons comprise a subset of R2 and R4 neurons. Error bars indicate SEM.