Abstract
Postzygotic reproductive isolation (RI) plays an important role in speciation. According to the stage at which it functions and the symptoms it displays, postzygotic RI can be called hybrid inviability, hybrid weakness or necrosis, hybrid sterility, or hybrid breakdown. In this review, we summarized new findings about hybrid incompatibilities in plants, most of which are from studies on Arabidopsis and rice. Recent progress suggests that hybrid incompatibility is a by-product of co-evolution either with “parasitic” selfish elements in the genome or with invasive microbes in the natural environment. We discuss the environmental influences on the expression of hybrid incompatibility and the possible effects of environment-dependent hybrid incompatibility on sympatric speciation. We also discuss the role of domestication on the evolution of hybrid incompatibilities.
Keywords: postzygotic reproductive barriers, Hybrid incompatibility, evolutionary force, Speciation genes, evolutionary genetics, Bateson-Dobzhansky–Muller model
Reproductive Isolation and Postzygotic Reproductive Barriers
Reproductive isolation (RI) hinders genetic exchange between species or populations. Therefore, it is considered fundamental to speciation (Coyne and Orr, 2004). Generally, RI in plants can be classified into prepollination and postpollination according to the developmental stage during which they occur. Prepollination isolations such as habitat divergence, temporal isolation, pollinator isolation, and mating system divergence usually function more effectively than postpollination isolations (Coyne and Orr, 2004). RI after postpollination can be divided into prezygotic isolation and postzygotic isolation. Postzygotic RI is common across the plant and animal kingdoms. Hybrids that undergo postzygotic RI are usually aborted or arrested after fertilization at different developmental stages or generations (Figure 1). According to the developmental stage at which postzygotic RI occurs and the symptoms it displays, postzygotic RI can be termed hybrid inviability, hybrid weakness or necrosis, hybrid sterility, or hybrid breakdown (Figure 1). In recent years, several genes involved in hybrid incompatibilities have been identified (Rieseberg and Blackman, 2010). These studies have extensively broadened our understanding of the molecular aspects of RI. In this review, we summarized new findings concerning the genetic and molecular control of intrinsic postzygotic hybrid incompatibility in plants and discuss the evolutionary basis of hybrid incompatibility in scenarios of evolution and domestication.
FIGURE 1.
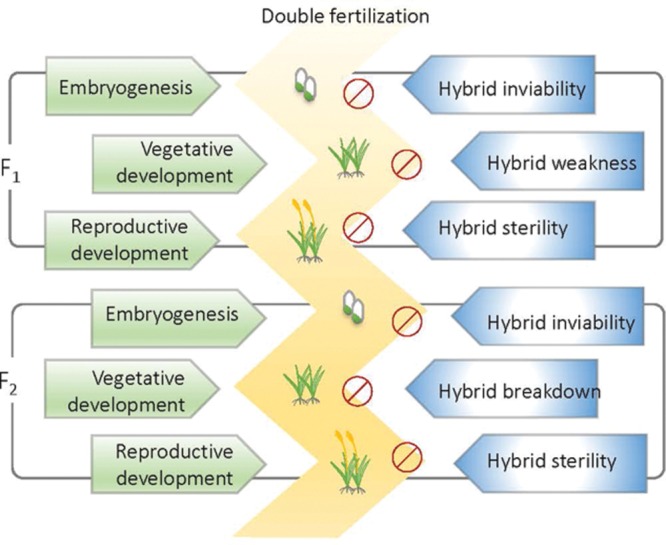
Different forms of postzygotic reproductive isolation (RI).
The Bateson-Dobzhansky–Muller (BDM) Model and Genetic Incompatibility
Because of its adverse effects, how hybrid incompatibility could be sustained in different species was an enigma to Darwin and his contemporaries. Fisher (1930) realized that geographical isolation is important for the establishment of hybrid incompatibility. In the early 20th century, Bateson, Dobzhansky, and Muller independently proposed the BDM model (Figure 2A) to give a genetic explanation of hybrid incompatibility (Coyne, 1992; Orr, 1996). The BDM model proposes that distinct variations accumulate in divergent populations. Some variations are not compatible with variations fixed in other populations although the loci or genes involved are compatible with their native genetic context. The deleterious effects of the incompatible variations cause hybrid failure and prevent gene flow (Figure 2). Therefore, genetic incompatibility is essential to the formation of postzygotic RI.
FIGURE 2.
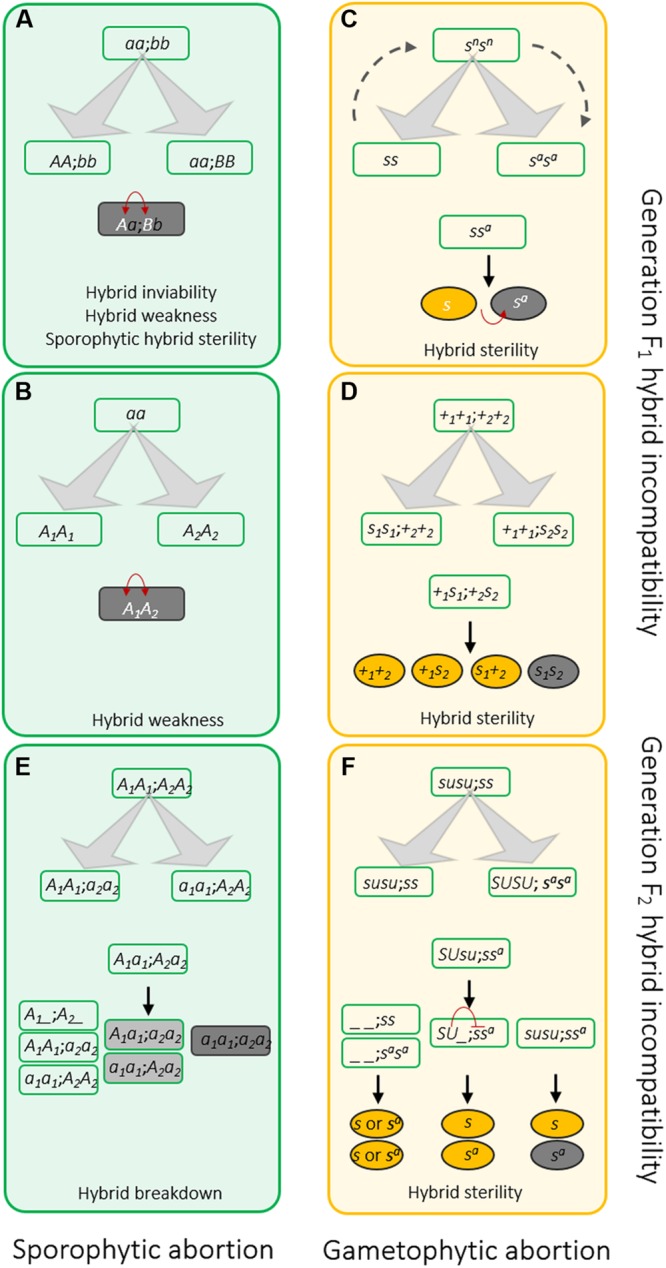
Genetic models of hybrid incompatibilities that occur in sporophytes (A,B,E) and gametophytes (C,D,F) at F1 generation (A–D) or later generation (E,F). (A) Classic two-locus interaction Bateson-Dobzhansky–Muller (BDM) model. Ancestral genes were independently mutated and sustained in parallel populations isolated by geographic barrier. The mutations are compatible to their own genetic contexts. However, mixture of these mutations are deleterious to the hybrids and leads to hybrid incompatibility. (B) The allelic interaction BDM model. Distinct from the two-locus involved BDM model, disharmonious interaction occurs between different alleles at one locus. (C) The sporo-gametophytic interaction model. Allelic interaction cause gamete abortion in the hybrids. The selfish nature of S allele can kill gametes with genotype Sa. The S and Sa alleles are likely evolved from a wild-compatible allele Sn, which is compatible with either S and Sa. No fertility issue can be observed in SSn and SaSn hybrids. Alternatively, Sn allele may also be the intermediate form of S and Sa during evolution. For example, after the ancestral allele S mutating to Sn, a second mutation of Sn produces Sa. (D) The duplicate gametic lethal model. At least one functional copy of the duplicated loci of S1 and S2 is required for gamete development. To avoid confusing of s1 and s2, the functional alleles (S1 and S2) are illustrated as +1 and +2 in the figure. (E) The duplicate recessive lethal model to elucidate genetic control of sporophytic F2 hybrid dysfunction. Duplicated A1 and A2 are essential to plants. Mutation at one locus is tolerated in plants due to the redundancy. However, if two populations carry mutations at different loci, their offspring with genotype a1a1a2a2 will show inferiority, owing to lack of any functional copy of A1 or A2. (F) A more complicated genetic model controlling F2 hybrid sterility identified in rice. During evolution, a regulator Su was evolved to suppress the selfish nature of the S allele described in (C) in a sporophytic manner. Only the ssa hybrids without SU showed sterility. The red arrows in (A–C) indicate deleterious genetic interactions between loci or alleles. The yellow and gray ovals in (D–F) indicate fertile and sterile pollens, respectively.
By comprehensively reviewing the literature, Presgraves (2010) suggested that selfish genetic elements such as repeat sequences, transposable elements, and meiotic drivers are likely to be the main cause of hybrid incompatibility. This conclusion is applicable to plants as well if one broadly considers mitochondria and chloroplasts to be selfish elements. Mitochondria and chloroplasts evolved from ancient bacterial symbionts, which are dependent on host reproduction for their transmission and therefore, can be considered as “reproductive parasites” (Werren, 2011). Nucleocytoplasmic incompatibility can usually be found to cause hybrid sterility in either interspecies or intraspecies hybridization. However, unlike that found in Drosophila (Bayes and Malik, 2009; Ferree and Barbash, 2009; Phadnis and Orr, 2009), the vast majority of well characterized postzygotic RIs are not directly correlated with selfish genetic elements in plants (Rieseberg and Blackman, 2010; Ouyang and Zhang, 2013). This suggests that the genetic control of postzygotic RI in plants may be distinct from that in animals.
Genetic and Molecular Regulation of Postzygotic RI
Disruption of Genomic Imprinting Associates with Hybrid Inviability
Hybrid inviability, or hybrid lethality, is common in higher plants and is a particularly strong barrier to interspecific gene flow compared with other hybrid incompatibility forms (Coyne and Orr, 2004). Hybrid seed failure is usually caused by developmental defects in the endosperm (Lafon-Placette and Köhler, 2016). The endosperm and embryo are products of double fertilization. The triploid endosperm is needed to nourish the developing diploid embryo (Olsen, 2001). Histological observations suggest that endosperm breakdown is always coupled with endosperm cellularization defects (Walia et al., 2009; Ishikawa et al., 2011). Embryo rescue is an effective way to rescue the hybrids (Sharmal et al., 1996), indicating that the endosperm is less tolerant of genetic incompatibility than the embryo.
To date, our understanding of hybrid inviability is very limited in terms of its molecular controls. Hybrid inviability is not usually a reciprocal postzygotic hybridization barrier (Lafon-Placette and Köhler, 2016). As an example, unidirectional hybrid inviability in rice was found between W593A (Oryza rufipogon) and T65wx (O. sativa ssp. japonica) when W593A was the female parent and T65wx was the male parent (Matsubara et al., 2003). A genetic study revealed that the incompatibility is controlled by the maternal nuclear gene Cif and the paternal nuclear gene cim (Matsubara et al., 2003). The parent-of-origin dependent effects of incompatible nuclear genes indicate that epigenetic conflicts, most likely genomic imprinting, may lead to hybrid inviability. Imprinted genes are parent-of-origin dependent and regulate endosperm development in plants (Köhler et al., 2012). Imprinting interruption was discovered in the endosperm of interspecific and intraspecific animal and plant hybrids (Vrana et al., 1998; Josefsson et al., 2006; Ishikawa et al., 2011; Wolf et al., 2014; Burkart-Waco et al., 2015; Kirkbride et al., 2015). Misregulation of some imprinted genes, such as MEDEA, FERTILIZATION-INDEPENDENT SEED 2, and the type-I MADS box genes PHERES1 and AGAMOUS-LIKE 62, has been associated with endosperm impairment in both interspecific and interploid Arabidopsis hybrids (Erilova et al., 2009; Walia et al., 2009; Jullien and Berger, 2010). Rice imprinted gene OsMADS87 may play a similar role in the regulation of endosperm development (Ishikawa et al., 2011; Chen et al., 2016). Moreover, suppression of certain imprinted genes can alleviate the low viability of interspecific and interploid seeds (Walia et al., 2009; Kradolfer et al., 2013a,b; Wolff et al., 2015). The maintenance of genomic imprinting involves complex interactions between trans-acting factors and regulatory cis-elements (Bartolomei and Ferguson-Smith, 2011). Normal endosperm development requires a highly specific balance of gene expression (Gutierrez-Marcos et al., 2003). Mismatch of trans- and cis-components and/or misregulation of the imprinted-gene-regulated genes between species may result in disruption of the balance and impair endosperm development, which eventually causes hybrid lethality. Examples of hybrid inviability controlled by multiple loci have been found in several plants (Chu and Oka, 1970; Burkart-Waco et al., 2012; Garner et al., 2015; Rebernig et al., 2015). Cloning of these genes will broaden our understanding of the molecular control of hybrid inviability.
Autoimmunity and Hybrid Weakness or Necrosis
Hybrid necrosis or weakness refers to the phenomena in which the hybrid shows developmental inferiority, such as necrotic leaves, small stature, or poor growth (Bomblies and Weigel, 2007). The weak hybrids often die before they reach the reproductive stage. Usually, the embryogenesis of plants expressing hybrid weakness is not affected (Saito et al., 2007; Chen et al., 2014). Classic genetic analysis indicated that deleterious BDM interactions between loci or alleles are the causes of hybrid necrosis and weakness (Figures 2A,B). In the past decade, several genes causing hybrid necrosis in plants have been cloned (Bomblies et al., 2007; Alcázar et al., 2009, 2010, 2014; Jeuken et al., 2009; Smith et al., 2011; Kradolfer et al., 2013a; Chen et al., 2014; Todesco et al., 2014), which comprehensively broaden our understandings of hybrid necrosis at molecular level. Almost all of the necrosis-causing genes identified from different plant species are likely to be associated with immune responses. In addition to the appearance of necrotic symptoms, pathogenesis-related genes are always activated (Bomblies et al., 2007; Alcázar et al., 2009; Chen et al., 2013). Though a few recessive genes have been found to induce the expression of hybrid necrosis (Bomblies and Weigel, 2007; Alcázar et al., 2009), most of the causal genes are dominant genes. This is consistent with the notion that most of the plant resistance genes are dominant genes. By systematically testing thousands of F1 hybrids, Chae et al. (2014) surprisingly found that there are autoimmune-caused hybrid necrosis hot spots in the Arabidopsis genome. Why the immune system is recruited in hybrid necrosis in various plant species is a fascinating but still open question. One reasonable explanation is that pathogens are a ubiquitous threat to plants. Microbe-driven selection accelerates the diversification of resistance genes (Jones and Dangl, 2006). The arm race between plants and pathogens facilitates the plant genome to accumulate more variations, making resistance genes one of the most diverse group of genes in plants (Bomblies, 2009). Due to fitness compensation, the immune system is delicately configured to ensure that defense responses will not be triggered unless invasion is detected. However, the diversity of defense-related genes substantially increases the risk of a defense regulation mismatch between different populations, which may activate defense responses in the absence of infection and cause hybrid necrosis and weakness as a fitness cost.
Gene Duplication Plays Vital Role in Hybrid Dysfunction
The duplicate recessive lethal model can be used to elucidate the genetic control of sporophytic F2 hybrid inviability and breakdown (Figure 2E). After a duplication event, there are two copies of gene A (A1 and A2) in the ancestor genome. Due to redundancy, mutations that accumulate in either gene copy can be sustained in the genome. It is assumed that mutation of A1 is kept in population 1 (a1a1A2A2), and mutation of A2 is sustained in population 2 (A1A1a2a2). A1a1A2a2 hybrids and the majority of the F2 progeny will be viable, except for the ones with genotype a1a1a2a2, since they lack a functional copy. An example is the HPA gene, which encodes histidinol-phosphate aminotransferase involving histidine biosynthesis and therefore, is essential for plant growth and development (Bikard et al., 2009). The Arabidopsis genome has two HPA genes that were generated through a small chromosome segmental duplication. The HPA copy on chromosome 1 has been shown to be completely silenced in Columbia-0 (Col), while the other copy on chromosome 5 is deleted from the genome of Cape Verde Island accession Cvi-0 (Cvi). Therefore, approximately one-sixteenth of the seeds (F2) produced by Col/Cvi F1 hybrids, which lack a functional HPA, are unable to maintain essential development, which leads to embryo arrest (Bikard et al., 2009). Hybrid chlorosis, another type of hybrid incompatibility, has been found in several plant species (Tomar and Singh, 1998; Ichitani et al., 2012; Nakano et al., 2015). Linkage analysis of the F2 hybrid chlorosis genes in rice found that the genes are located on the distal regions of the short arms of chromosomes 12 and 11, respectively (Ichitani et al., 2012). Previous study showed that these regions are very conserved and were produced by chromosomal duplication (Wu et al., 1998). Therefore, this study may offer another case of disruption of duplicate genes causing hybrid incompatibility.
Oka (1988) conceived a similar model to explain the two loci-controlled F1 gametophytic sterility in rice. According to this model, two duplicated loci (S1 and S2) are important for gamete development. At least one dominant allele of these two loci (+1 or +2) is required. Therefore, 25% of the gametes produced by the hybrid (S1s1/S2s2), the ones with genotype s1s2, will be aborted due to a lack of +1 or +2. Recently, studies in rice provided molecular support for this model (Mizuta et al., 2010; Yamagata et al., 2010), which emphasizes a role of genetic drift in the evolution of hybrid incompatibility. As an example, a recent small-scale gene duplication event occurring after Oryza–Brachypodium differentiation was found to involve in the hybrid sterility of intraspecific rice hybrids (Mizuta et al., 2010). The disruption of duplicated gene DOPPELGANGER1 (DPL1) in japonica rice cultivar Niponbare and DPL2 in indica rice cultivar Kasalash causes pollen sterility in F1 hybrids (Mizuta et al., 2010). These findings confirm that gene duplication is important to hybrid incompatibility events and possibly to speciation.
Complicated Genetic-Interaction-Networks Involve in Hybrid Sterility
Hybrid sterility is the most prevalent form of postzygotic RI in the plant and animal kingdoms (Ouyang et al., 2010; Rieseberg and Blackman, 2010; Maheshwari and Barbash, 2011). The hybrids develop vitally at the vegetative stage, whereas sterility appears in either the F1 or F2 generation. From a cytological perspective, fertility defects could be caused by degradation either of the generative cells (n), including pollen, the embryo-sac, or, in some cases, both, or of the vegetative/maternal tissues (2n) that enclose or surround the gametes, for instance, the tapetum cells. From this point of view, hybrid sterility could be sporophytic or gametophytic. Hybrid fertility determines the appearance of heterosis. Due to its practical importance, the genetic control of hybrid sterility has been comprehensively studied in crops. After six decades of study, more than 50 rice loci governing hybrid infertility have been identified to date (Kubo, 2013). Genetically, disharmonious allelic interaction is the major cause of F1 hybrid sterility, though sometimes, the deleterious interaction may involve two loci, which can be explained by Oka’s duplicate gametic lethal model aforementioned (Figure 2D).
The sporo-gametophytic interaction model (Figure 2C) proposed by Kitamura is a popular way to illustrate the allelic interactions that cause gametophytic sterility (Oka, 1988). The hybrid sterile gene (S) functions against its opposite allele (Sa) in the hybrids (S/Sa) during gametogenesis. Due to the selfish nature of S, the gametes carrying Sa will be killed through an as-yet unidentified mechanism. Recently, a few of such hybrid sterility genes have been cloned and functionally characterized. The genes S5 and Sa condition female and male sterility, respectively, in indica-japonica hybrids (Chen et al., 2008; Long et al., 2008; Yang et al., 2012). Initially, an aspartic protease-encoding gene that displays a two nucleotide difference between the japonica allele (S5j) and indica allele (S5i) was thought to be responsible for the incompatibility (Chen et al., 2008). Further study showed that two adjacent genes are also required to induce hybrid sterility (Yang et al., 2012). The three genes constitute a killer-protector system through switching on/off ER stress-induced premature programmed cell death in the embryo-sac, which determines megaspore fertility (Yang et al., 2012). Interestingly, the Sa locus also includes two adjacent genes, which encode a small ubiquitin-like modifier E3 ligase-like protein and an F-box protein, respectively (Long et al., 2008). In vitro experiments demonstrated that the products of the two causal genes can physically interact (Long et al., 2008). However, the underlying mechanism remains to be elucidated. These studies strongly suggest that this kind of pseudo-allelic interaction-controlled hybrid sterility may be more complicated than the sporo-gametophytic interaction model implies. In some cases, there exists a third allele, Sn, that is compatible with either S or Sa without a fertility issue in the hybrids (Oka, 1988). From an evolutionary perspective, Sn may be the bridge between S and Sa during evolutionary divergence. Sn could either be the ancestral form of parallelly evolved S and Sa or it may be the intermediate between sequentially evolved S and Sa. Intriguingly, some gamete eliminators can be inactivated by an unlinked suppressor (Kubo et al., 2011). Such a suppressor, together with the gamete eliminator, could be employed as a neutral allele for evolution (Figure 2F).
F2 hybrid sterility is observed in various plant species (Kubo and Yoshimura, 2005; Sweigart et al., 2006; Yi et al., 2006; Li et al., 2015). Unlike F1 sterility, gamete abortion can be observed in some F2 individuals derived from a completely fertile F1 hybrid. Although the duplicate recessive lethal model (Figure 2E) can be applied to some sporophytic F2 hybrid sterility cases (Yi et al., 2010), recent studies in rice imply that the genetic control of F2 hybrid sterility can be more complicated (Figure 2F). As an example, S24 was initially identified from chromosome segment substitution lines and acts gametophytically. In backcrossed F1, pollens carrying japonica allele S24 (S24j) are killed due to the selfish nature of the indica allele (S24i) (Kubo et al., 2008). Nevertheless, further analysis revealed that the deleterious effect of S24i is also suppressed by another dominant sporophytic gene, ESF, from indica (Kubo et al., 2011). Therefore, S24i is unable to eliminate S24j in indica/japonica F1 hybrids because of the existence of ESF. However, in the segregated F2 population, S24 is released from ESF in some progeny of the esf genotype, and this causes sterility at the F2 generation. Recently, another independent genetic pathway mediated by S35 and INK was identified from the same parental derived population, indicating complicated genetic regulation of hybrid sterility (Kubo et al., 2016b). Interestingly, a recent study found that an initially reported F2 hybrid sterility causal locus hsa1 of rice is composed by two tightly linked genes, and this locus may involve either F1 or F2 hybrid sterility events (Kubo et al., 2016a). This study identified a potential link between F1 and F2 sterility based on some common genetic elements. However, the combination of different elements in various germplasm may determine the genetic action manner of the genes.
Cytoplasmic male sterility (CMS) is another widely distributed type of hybrid sterility in many organisms. Plant CMS is determined by abnormal mitochondrial genes, which usually are gain-of-function open reading frames (ORFs) comprising segments derived from mitochondrial gene-coding and gene-flanking sequences and cause pollen failure of plants (reviewed in Chase, 2007). Many of these CMS mitochondrial genes can be suppressed or counteracted by the products of one or more nuclear encoded restorer genes. Several restorer genes have been identified in various plant species, most of which encoding pentatricopeptide-repeat proteins (PPRs; Bentolila et al., 2002; Brown et al., 2003; Desloire et al., 2003; Komori et al., 2004; Klein et al., 2005; Uyttewaal et al., 2008; Itabashi et al., 2011; Hu et al., 2012; Huang et al., 2015). These PPRs can suppress the accumulation of abnormal CMS mitochondrial transcripts (reviewed in Chase, 2007). Because mitochondria are usually maternally inherited, CMS is typically transmitted through female gametes, while restorer genes are transmitted through both male and female gametes. Mismatch or lack of restorer genes for the mitochondrial CMS gene in the hybrids will lead to hybrid sterility.
Evolution of Hybrid Incompatibilities
An overview of the characterized speciation genes has shown that, in many cases, the evolution of hybrid incompatibility is likely not caused by adaptive mutations, but by arm races between selfish genetic elements and the host genes that regulate or suppress them (Presgraves, 2010). Notably, hybrid sterility is predominantly affected by selfish elements in animals (Presgraves, 2010; Maheshwari and Barbash, 2011). Selfish element-driven hybrid incompatibility is also prevalent in plant species and is responsible for the evolution of CMS (Rieseberg and Blackman, 2010). However, how selfish genetic elements can become fixed in a population remains to be elucidated because these elements, obviously, are usually detrimental to the host genome.
Several studies of plants have suggested that some genes responsible for hybrid necrosis and weakness are likely maintained by selection (Alcázar et al., 2010; Chen et al., 2014; Sicard et al., 2015). For instance, a rare allele of the Arabidopsis Strubbelig Receptor Family 3 gene shows a recent selective sweep signature in Central Asian populations (Alcázar et al., 2010). Balancing selection was implied to act in the maintaining of the diversity of some hybrid necrosis causal genes in different species (Chen et al., 2014; Sicard et al., 2015). A recent study suggested that the hms2 interacting BDM locus, hms1, of Mimulus causing hybrid sterility shows strong natural selection signature (Sweigart and Flagel, 2015). In addition, a gene responsible for incompatibility can be indirectly selected if the gene is physically adjacent to an adaptive gene (Presgraves, 2013). As an example, a population of yellow monkey flower has recently evolved an adaption to the tailings of local copper mines (Wright et al., 2013). When it crosses with off-mine plants, hybrid necrosis syndromes show an association with the copper tolerance. Recently, it was found that the necrosis locus Nec1 is genetically linked with the tolerance locus Tol1 (Wright et al., 2013). Hence, the selection pressure imposed by Tol1 would help sustain Nec1 in the mine-adapted population. Likewise, crop domestication may also increase the likelihood of genes causing incompatibility to fix in the population through genetic hitchhiking. For example, rice gamete eliminator S5 is linked with OsC1, a gene targeted during domestication to control the color of the leaf sheath and apiculus (Saitoh et al., 2004).
These findings suggest that hybrid incompatibility is a by-product of co-evolution in the arm races between host and internal “parasitic” selfish elements or invasive microbes in the external environment. Different species have evolved distinct strategies to prevent invasion. Mismatch of regulation machineries is the cause of hybrid incompatibility per se. However, once a barrier is established, is it possible that selection directly favors the hybrid incompatibilities? Hybrid lethality is definitely disadvantageous. Nevertheless, the plants can sometimes derive benefit from it. If the hybrids are vigorous at the vegetative stage but are sterile or show weakness in the next generation, they occupy space and resources to compete with their parents. Under this circumstance, reproductive barriers that occur at an earlier stage may be selected to avoid producing inferior hybrids at later stages or the next generation. This allows the effective use of limited resources. For example, outbreeding species O. longistaminata often inhabits sympatrically with O. breviligulata and O. glaberrima (Oka, 1988). Hybrids between O. longistaminata and O. breviligulata or O. glaberrima usually suffer from hybrid inviability (Chu and Oka, 1970). A few seeds can occasionally be produced, but they are always weak and semisterile (Oka, 1988). Therefore, the complementary incompatible genes inducing hybrid inviability can be regionally increased to prevent resource consumption by the weak and sterile F1 plants. This is a possible explanation for why hybrid inviability is prevalent in wild rice of African origin.
Environment and Hybrid Incompatibilities
Environmentally dependent genetic incompatibilities have been found in different species. By surveying the RI between 27 naturally compatible yeast isolates under 20 distinct environmental conditions, Hou et al. (2015) found that environment-dependent hybrid incompatibility is not rare. This conditionally expressed RI is also observed in plants. Most of the hybrid necrosis or weakness cases that have been studied so far are temperature sensitive (Bomblies et al., 2007; Saito et al., 2007; Alcázar et al., 2009; Fu et al., 2013; Chen et al., 2014). Usually, low temperature promotes the expression of inferior symptoms. This is consistent with the notion that high temperature can inhibit plant immunity (Traw and Bergelson, 2010; Alcázar and Parker, 2011; Hua, 2013). As an exceptional, contrasting case found in rice, Hwi1/Hwi2-induced hybrid weakness is suppressed by low temperature (Chen et al., 2014). For this case, the causal locus Hwi1 consists of two indispensable LRR-RLK homologs, while its incompatible gene Hwi2 encodes a secreted protease. This two-locus/three-gene system was assumed to overactivate defense responses through pattern-triggered immunity (PTI). A very interesting study recently showed that plants preferentially activate effector-triggered immunity (ETI) at low temperatures and PTI at high temperatures, possibly indicating that the genes involved in ETI are more likely to be recruited for establishing hybrid incompatibility between species (Cheng et al., 2013). Hybrid incompatibilities can also be affected by temperature in animals (Barbash et al., 2000; Koevoets et al., 2012). These examples suggest environmental factors such as temperature may interact with genetic control to determine the expression of hybrid incompatibilities.
Geographical separation plays a vital role in speciation (Coyne and Orr, 2004). This is supported by the notion that most previously studied hybrid necrosis cases involve rare alleles found in geographically unrelated populations. However, at least one case found in Arabidopsis showed that the alleles involved co-exist in a certain place at high frequency, which means inferior hybrids can be observed in these populations in the wild (Todesco et al., 2014). As an explanation, the inferior hybrids may have a conditional advantage to maintain all the alleles in the population. However, once the adverse environment fails to persist, the alleles involved may contribute to sympatric speciation. Take hybrid necrosis as an example; populations inhabiting in the same niche carry different resistance genes that are maintained by selection. Initially, the local temperature is above the threshold to trigger the inferior symptoms such as necrosis. Thus, the populations are completely compatible. However, when the local climate changes and the temperature decreases, the previously compatible genes become incompatible and hinder genetic flow between the populations (Figure 3). Next, we need more evidences to support this hypothesis.
FIGURE 3.
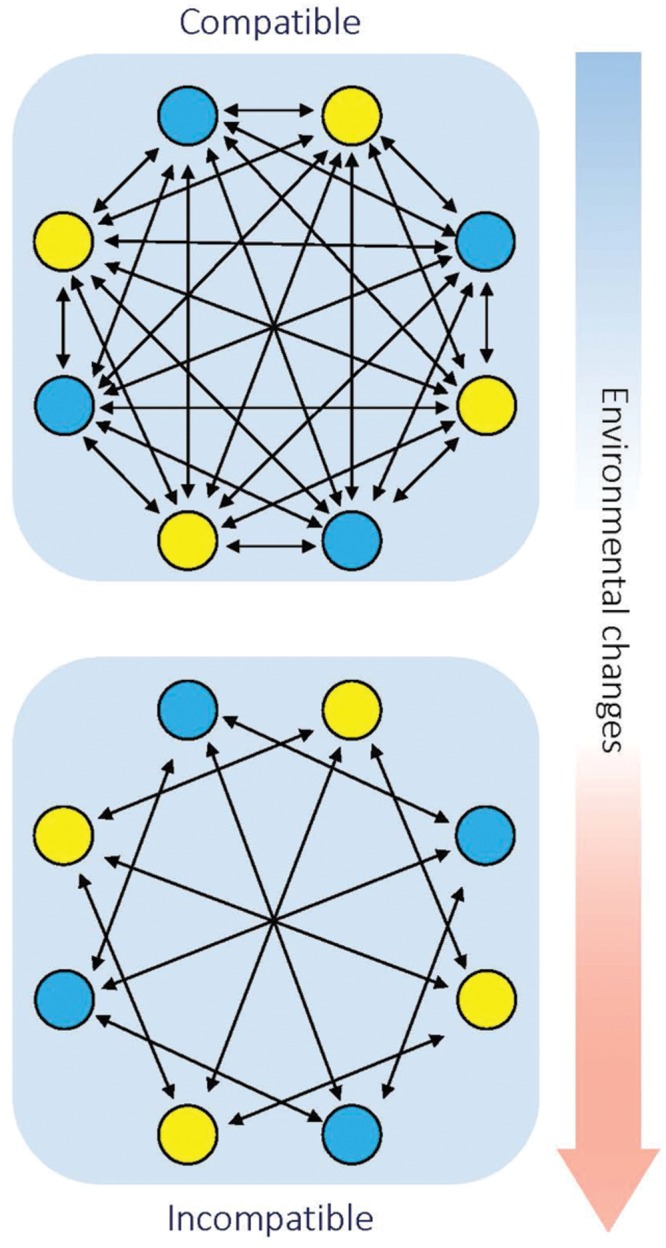
Diagram of how environmentally conditioned hybrid incompatibility contributes to sympatric speciation. Arrows indicate gene flow between individuals. Blue and yellow dots indicate individuals with different genotypes that inhabit the same region. Initially, individuals with different genotype are compatible with each other. Along with environmental changes (such as decreasing temperature), environmentally conditioned genetic incompatibility leads to hybrid incompatibility between different genotypes.
Domestication and Crop Improvement May Contribute to Hybrid Incompatibility
Hybrid incompatibility is a by-product of evolution (Coyne and Orr, 2004). In terms of crops, artificial selection and hybridization accelerate the evolutionary process (Meyer and Purugganan, 2013). A literature survey indicated that majority of the most economically important crops were isolated from their progenitors with the existence or evolution of prezygotic and/or postzygotic reproductive barriers, even though geographic isolation was absent during domestication, at least in the initial stages of domestication for most species (Dempewolf et al., 2012). More important to early human farmers, RI can facilitate the maintenance of gene combinations during domestication. By taking these into account, Dempewolf et al. (2012) raised the question whether RI could be viewed as a long-overlooked “domestication trait,” though this hypothesis requires more supporting evidence.
To date, a few genes of hybrid incompatibility in crops have been isolated, however, the evolutionary study of these genes are very limited. Among them, S5, one of the key determinants of japonica-indica sterility, is about 400 kb apart from OsC1, which is responsible for anthocyanin pigmentation of rice. Loss-of-function OsC1 alleles were selected during domestication (Saitoh et al., 2004). Molecular population analysis revealed that japonica and indica rice have independent origins of OsC1 (Saitoh et al., 2004). Interestingly, japonica and indica S5 alleles were differentiated before domestication (Yang et al., 2012). It is unknown whether japonica and indica specific OsC1 alleles were co-fixed with different S5 alleles in rice cultivars during domestication. However, it is reasonable to assume that the selection of OsC1 imposed an indirect force to sustain different S5 alleles in different rice subspecies. Similar situation is observed at Sa locus, which located within a genomic region with strong selective sweep signature in japonica as well as indica rice (Huang et al., 2012). Yield is a key trait for crop domestication and improvement. Gn1a is important for rice yield formation (Ashikari et al., 2005). It is found that artificial selection of Gn1a plays an important role in improving rice yields across different ecological regions (Wang et al., 2015). Notably, Gn1a is linked with S35 that determines pollen sterility of japonica-indica hybrids (Kubo, 2013). It will be interesting to interpret how the artificial selection of Gn1a functions on S35 during domestication. Breeding can also affect the distribution of incompatible genes in crops. An unambiguous example comes from the geographical study of wheat hybrid necrosis inducing gene Ne2. Due to genetic linkage with the brown rust resistance gene, the frequency of Ne2 in modern varieties has risen considerably in the past 100 years (Pukhalskiy et al., 2000). Moreover, at least one example of tomato showed that the gene involves in hybrid incompatibility may be directly selected by breeders for crop improvement. A Lycopersicon pimpinellifolium gene Cf-2 confers resistance to the fungus Cladosporium Fulvum in an Rcr3 dependent manner (Krüger et al., 2002). Rcr3 was maintained by balancing selection and co-evolved with Cf-2 in wild tomato species (Hörger et al., 2012). Cf-2 has been bred into cultivated tomato (L. esculentum) for resistance improvement. However, Cf-2 of wild tomato and Rcr3 of cultivated tomato are able to interact with each other to induce hybrid necrosis syndrome in the hybrids (Krüger et al., 2002).
Collectively, genetic hitchhiking effect and direct selection of the hybrid incompatible genes can contribute to the RI in the history of crop domestication and improvement. However, we should note that all the hybrid incompatible genes are pre-existing in nature, which means domestication and breeding programs may comprehensively change the distribution and sustention of hybrid incompatible genes, rather than create new ones. Cloning and evolutionary analysis of the RI causal genes may provide novel insights to our understanding of how domestication affects RI and speciation. Rice, as a model organism for plant sciences, is one of the most suitable crops to test this hypothesis. Asian cultivated rice has two subspecies, japonica and indica, that are not fully isolated and can be regarded as the early stage of species divergence. African cultivated rice was domesticated independently of Asian rice. RI is very common between these species, including their progenitors (Chu and Oka, 1970; Oka, 1988). Several loci responsible for the interspecies or intraspecies hybrid incompatibilities have been mapped on different chromosomes. Cloning these hybrid incompatibility genes and studying their molecular and biochemical control in rice, as well as other plants, will substantially help us understand the evolution of RI.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Priority Academic Development of Jiangsu Higher Education Institutions, Natural Science Foundation of Jiangsu Province (BK20150446) and National Natural Science Foundation of China (31571623).
References
- Alcázar R., García A. V., Kronholm I., de Meaux J., Koornneef M., Parker J. E., et al. (2010). Natural variation at strubbelig receptor kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat. Genet. 42 1135–1139. 10.1038/ng.704 [DOI] [PubMed] [Google Scholar]
- Alcázar R., García A. V., Parker J. E., Reymond M. (2009). Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc. Natl. Acad. Sci. U.S.A. 106 334–339. 10.1073/pnas.0811734106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcázar R., Parker J. E. (2011). The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16 666–675. 10.1016/j.tplants.2011.09.001 [DOI] [PubMed] [Google Scholar]
- Alcázar R., von Reth M., Bautor J., Chae E., Weigel D., Koornneef M., et al. (2014). Analysis of a plant complex resistance gene locus underlying immune-related hybrid incompatibility and its occurrence in nature. PLoS Genet. 10:e1004848 10.1371/journal.pgen.1004848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309 741–745. 10.1126/science.1113373 [DOI] [PubMed] [Google Scholar]
- Barbash D. A., Roote J., Ashburner M. (2000). The Drosophila melanogaster hybrid male rescue gene causes inviability in male and female species hybrids. Genetics 154 1747–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M. S., Ferguson-Smith A. C. (2011). Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 3:a002592 10.1101/cshperspect.a002592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes J. J., Malik H. S. (2009). Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326 1538–1541. 10.1126/science.1181756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S., Alfonso A. A., Hanson M. R. (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. U.S.A. 99 10887–10892. 10.1073/pnas.102301599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Patel D., Le Metté C., Giorgi V., Camilleri C., Bennett M. J., et al. (2009). Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323 623–626. 10.1126/science.1165917 [DOI] [PubMed] [Google Scholar]
- Bomblies K. (2009). Too much of a good thing? Hybrid necrosis as a by-product of plant immune system diversification. Botany 87 1013–1022. [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., Dangl J. L., et al. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5:e236 10.1371/journal.pbio.0050236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Weigel D. (2007). Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8 382–393. 10.1038/nrg2082 [DOI] [PubMed] [Google Scholar]
- Brown G. G., Formanová N., Jin H., Wargachuk R., Dendy C., Patil P., et al. (2003). The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35 262–272. 10.1046/j.1365-313X.2003.01799.x [DOI] [PubMed] [Google Scholar]
- Burkart-Waco D., Josefsson C., Dilkes B., Kozloff N., Torjek O., Meyer R., et al. (2012). Hybrid incompatibility in Arabidopsis is determined by a multiple-locus genetic network. Plant Physiol. 158 801–812. 10.1104/pp.111.188706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart-Waco D., Ngo K., Lieberman M., Comai L. (2015). Perturbation of parentally biased gene expression during interspecific hybridization. PLoS ONE 10:e0117293 10.1371/journal.pone.0117293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E., Bomblies K., Kim S. T., Karelina D., Zaidem M., Ossowski S., et al. (2014). Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 156 1341–1351. 10.1016/j.cell.2014.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase C. D. (2007). Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23 81–90. 10.1016/j.tig.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Chen C., Begcy K., Liu K., Folsom J. J., Wang Z., Zhang C., et al. (2016). Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol. 171 606–622. 10.1104/pp.15.01992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Lin Y. S., Shen J. B., Shan J. X., Qi P., et al. (2014). A two-locus interaction causes interspecific hybrid weakness in rice. Nat. Commun. 5:3357 10.1038/ncomms4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Shan J. X., Zhu M. Z., Shi M., Gao J. P., et al. (2013). Genetic and physiological analysis of a novel type of interspecific hybrid weakness in rice. Mol. Plant 6 716–728. 10.1093/mp/sss146 [DOI] [PubMed] [Google Scholar]
- Chen J., Ding J., Ouyang Y., Du H., Yang J., Cheng K., et al. (2008). A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc. Natl. Acad. Sci. U.S.A. 105 11436–11441. 10.1073/pnas.0804761105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Gao X., Feng B., Sheen J., Shan L., He P. (2013). Plant immune response to pathogens differs with changing temperatures. Nat. Commun. 4:2530 10.1038/ncomms3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y., Oka H. (1970). The genetic basis of crossing barriers between Oryza perennis subsp. barthii and its related taxa. Evolution 24 135–144. [DOI] [PubMed] [Google Scholar]
- Coyne J. A. (1992). Genetics and speciation. Nature 355 511–515. 10.1038/355511a0 [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A. (2004). Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Dempewolf H., Hodgins K. A., Rummell S. E., Ellstrand N. C., Rieseberg L. H. (2012). Reproductive isolation during domestication. Plant Cell 24 2710–2717. 10.1105/tpc.112.100115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desloire S., Gherbi H., Laloui W., Marhadour S., Clouet V., Cattolico L., et al. (2003). Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erilova A., Brownfield L., Exner V., Rosa M., Twell D., Scheid O. M., et al. (2009). Imprinting of the Polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 5:e1000663 10.1371/journal.pgen.1000663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree P. M., Barbash D. A. (2009). Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7:e1000234 10.1371/journal.pbio.1000234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F. (1930). The Genetical Theory of Natural Selection. Oxford: Oxford University Press. [Google Scholar]
- Fu C. Y., Wang F., Sun B. R., Liu W. G., Li J. H., Deng R. F., et al. (2013). Genetic and cytological analysis of a novel type of low temperature-dependent intrasubspecific hybrid weakness in rice. PLoS ONE 8:e73886 10.1371/journal.pone.0073886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner A. G., Kenney A. M., Fishman L., Sweigart A. L. (2015). Genetic loci with parent-of-origin effects cause hybrid seed lethality between Mimulus species. New Phytol. 211 319–331. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos J. F., Pennington P. D., Costa L. M., Dickinson H. G. (2003). Imprinting in the endosperm: a possible role in preventing wide hybridization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 1105–1111. 10.1098/rstb.2003.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörger A. C., Ilyas M., Stephan W., Tellier A., van der Hoorn R. A. L., Rose L. E. (2012). Balancing selection at the tomato RCR3 guardee gene family maintains variation in strength of pathogen defense. PLoS Genet. 8:e1002813 10.1371/journal.pgen.1002813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Friedrich A., Gounot J.-S., Schacherer J. (2015). Comprehensive survey of condition-specific reproductive isolation reveals genetic incompatibility in yeast. Nat. Commun. 6:7214 10.1038/ncomms8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wang K., Huang W., Liu G., Gao Y., Wang J., et al. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24 109–122. 10.1105/tpc.111.093211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J. (2013). Modulation of plant immunity by light, circadian rhythm, and temperature. Curr. Opin. Plant Biol. 16 406–413. 10.1016/j.pbi.2013.06.017 [DOI] [PubMed] [Google Scholar]
- Huang W., Yu C., Hu J., Wang L., Dan Z., Zhou W., et al. (2015). Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility. Proc. Natl. Acad. Sci. U.S.A. 112 14984–14989. 10.1073/pnas.1511748112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Kurata N., Wei X., Wang Z. X., Wang A., Zhao Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490 497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichitani K., Takemoto Y., Iiyama K., Taura S., Sato M. (2012). Chromosomal location of HCA1 and HCA2 hybrid chlorosis genes in rice. Int. J. Plant Genomics 2012:6490811 10.1155/2012/649081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R., Ohnishi T., Kinoshita Y., Eiguchi M., Kurata N., Kinoshita T. (2011). Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J. 65 798–806. 10.1111/j.1365-313X.2010.04466.x [DOI] [PubMed] [Google Scholar]
- Itabashi E., Iwata N., Fujii S., Kazama T., Toriyama K. (2011). The fertility restorer gene, Rf2 for lead rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. Plant J. 65 359–367. 10.1111/j.1365-313X.2010.04427.x [DOI] [PubMed] [Google Scholar]
- Jeuken M. J. W., Zhang N. W., McHale L. K., Pelgrom K., den Boer E., Lindhout P., et al. (2009). Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21 3368–3378. 10.1105/tpc.109.070334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Josefsson C., Dilkes B., Comai L. (2006). Parent-dependent loss of gene silencing during interspecies hybridization. Curr. Biol. 16 1322–1328. 10.1016/j.cub.2006.05.045 [DOI] [PubMed] [Google Scholar]
- Jullien P. E., Berger F. (2010). Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet. 6:e1000885 10.1371/journal.pgen.1000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkbride R. C., Yu H. H., Nah G., Zhang C., Shi X., Chen Z. J. (2015). An epigenetic role for disrupted paternal gene expression in postzygotic seed abortion in Arabidopsis interspecific hybrids. Mol. Plant 8 1766–1775. 10.1016/j.molp.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Klein R. R., Klein P. E., Mullet J. E., Minx P., Rooney W. L., Schertz K. F. (2005). Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor. Appl. Genet. 111 994–1012. 10.1007/s00122-005-2011-y [DOI] [PubMed] [Google Scholar]
- Koevoets T., Van De Zande L., Beukeboom L. W. (2012). Temperature stress increases hybrid incompatibilities in the parasitic wasp genus Nasonia. J. Evol. Biol. 25 304–316. 10.1111/j.1420-9101.2011.02424.x [DOI] [PubMed] [Google Scholar]
- Köhler C., Wolff P., Spillane C. (2012). Epigenetic mechanisms underlying genomic imprinting in plants. Annu. Rev. Plant Biol. 63 331–352. 10.1146/annurev-arplant-042811-105514 [DOI] [PubMed] [Google Scholar]
- Komori T., Ohta S., Murai N., Takakura Y., Kuraya Y., Suzuki S., et al. (2004). Map-based cloning of a fertility restorer gene, Rf-1 in rice (Oryza sativa L.). Plant J. 37 315–325. [DOI] [PubMed] [Google Scholar]
- Kradolfer D., Hennig L., Köhler C. (2013a). Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development. PLoS Genet. 9:e1003163 10.1371/journal.pgen.1003163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kradolfer D., Wolff P., Jiang H., Siretskiy A., Kohler C. (2013b). An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev. Cell 26 525–535. 10.1016/j.devcel.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Krüger J., Thomas C. M., Golstein C., Dixon M. S., Smoker M., Tang S., et al. (2002). A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296 744–747. 10.1126/science.1069288 [DOI] [PubMed] [Google Scholar]
- Kubo T. (2013). Genetic mechanisms of postzygotic reproductive isolation: Aan epistatic network in rice. Breed. Sci. 63 359–366. 10.1270/jsbbs.63.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Takashi T., Ashikari M., Yoshimura A., Kurata N. (2016a). Two tightly linked genes at the hsa1 locus cause both F1 and F2 hybrid sterility in rice. Mol. Plant 9 221–232. 10.1016/j.molp.2015.09.014 [DOI] [PubMed] [Google Scholar]
- Kubo T., Yamagata Y., Eguchi M., Yoshimura A. (2008). A novel epistatic interaction at two loci causing hybrid male sterility in an inter-subspecific cross of rice (Oryza sativa L.). Genes Genet. Syst. 83 443–453. 10.1266/ggs.83.443 [DOI] [PubMed] [Google Scholar]
- Kubo T., Yoshimura A. (2005). Epistasis underlying female sterility detected in hybrid breakdown in a japonica-indica cross of rice (Oryza sativa L.). Theor. Appl. Genet. 110 346–355. 10.1007/s00122-004-1846-y [DOI] [PubMed] [Google Scholar]
- Kubo T., Yoshimura A., Kurata N. (2011). Hybrid male sterility in rice is due to epistatic interactions with a pollen killer locus. Genetics 189 1083–1092. 10.1534/genetics.111.132035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Yoshimura A., Kurata N. (2016b). Pollen killer gene S35 function requires interaction with an activator that maps close to S24., another pollen killer gene in rice. G3 6 1459–1468. 10.1534/g3.116.027573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C., Köhler C. (2016). Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Mol. Ecol. 25 2620–2629. 10.1111/mec.13552 [DOI] [PubMed] [Google Scholar]
- Li R., Guo M., Lu Y., Yang Y., Liu M., Zhu Q., et al. (2015). Genetic dissection of hybrid breakdown in an indica/japonica cross and fine mapping of a quantitative trait locus qSF-12 in rice (Oryza sativa L.). Mol. Breed. 35 1–12. 10.1007/s11032-015-0331-4 [DOI] [Google Scholar]
- Long Y., Zhao L., Niu B., Su J., Wu H., Chen Y., et al. (2008). Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. U.S.A. 105 18871–18876. 10.1073/pnas.0810108105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S., Barbash D. A. (2011). The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45 331–355. 10.1146/annurev-genet-110410-132514 [DOI] [PubMed] [Google Scholar]
- Matsubara K., Khin-Thidar, Sano Y. (2003). A gene block causing cross-incompatibility hidden in wild and cultivated rice. Genetics 165 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. S., Purugganan M. D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14 840–852. 10.1038/nrg3605 [DOI] [PubMed] [Google Scholar]
- Mizuta Y., Harushima Y., Kurata N. (2010). Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. U.S.A. 107 20417–20422. 10.1073/pnas.1003124107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Mizuno N., Tosa Y., Yoshida K., Park P., Takumi S. (2015). Accelerated senescence and enhanced disease resistance in hybrid chlorosis lines derived from interspecific crosses between tetraploid wheat and Aegilops tauschii. PLoS ONE 10:e0121583 10.1371/journal.pone.0121583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka H.-I. (ed.) (1988). Origin of Cultivated Rice. Amsterdam: Elsevier. [Google Scholar]
- Olsen O.-A. (2001). ENDOSPERM DEVELOPMENT: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 233–267. 10.1146/annurev.arplant.52.1.233 [DOI] [PubMed] [Google Scholar]
- Orr H. A. (1996). Dobzhansky, Bateson, and the genetics of speciation. Genetics 144 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y., Liu Y. G., Zhang Q. (2010). Hybrid sterility in plant: stories from rice. Curr. Opin. Plant Biol. 13 186–192. 10.1016/j.pbi.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Ouyang Y., Zhang Q. (2013). Understanding reproductive isolation based on the rice model. Annu. Rev. Plant Biol. 64 111–135. 10.1146/annurev-arplant-050312-120205 [DOI] [PubMed] [Google Scholar]
- Phadnis N., Orr H. A. (2009). A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323 376–379. 10.1126/science.1163934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C. (2010). The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11 175–180. 10.1038/nrg2718 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C. (2013). Hitchhiking to speciation. PLoS Biol. 11:e1001498 10.1371/journal.pbio.1001498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukhalskiy V. A., Martynov S. P., Dobrotvorskaya T. V. (2000). Analysis of geographical and breeding-related distribution of hybrid necrosis genes in bread wheat (Triticum aestivum L.). Euphytica 114 233–240. 10.1023/A:1003915708448 [DOI] [Google Scholar]
- Rebernig C. A., Lafon-Placette C., Hatorangan M. R., Slotte T., Köhler C. (2015). Non-reciprocal interspecies hybridization barriers in the Capsella genus are established in the Endosperm. PLoS Genet. 11:e1005295 10.1371/journal.pgen.1005295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Blackman B. K. (2010). Speciation genes in plants. Ann. Bot. 106 439–455. 10.1093/aob/mcq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Ichitani K., Suzuki T., Marubashi W., Kuboyama T. (2007). Developmental observation and high temperature rescue from hybrid weakness in a cross between Japanese rice cultivars and Peruvian rice cultivar “Jamaica.”. Breed. Sci. 57 281–288. 10.1270/jsbbs.57.281 [DOI] [Google Scholar]
- Saitoh K., Onishi K., Mikami I., Thidar K., Sano Y. (2004). Allelic diversification at the C (OsC1) locus of wild and cultivated rice: Nnucleotide changes associated with phenotypes. Genetics 168 997–1007. 10.1534/genetics.103.018390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmal D. R., Kaur R., Kumar K. (1996). Embryo rescue in plants – a review. Euphytica 89 325–337. [Google Scholar]
- Sicard A., Kappel C., Josephs E. B., Lee Y. W., Marona C., Stinchcombe J. R., et al. (2015). Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in Capsella. Nat. Commun. 6:7960 10.1038/ncomms8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M., Bomblies K., Weigel D. (2011). Complex evolutionary events at a tandem cluster of Arabidopsis thaliana genes resulting in a single-locus genetic incompatibility. PLoS Genet. 7:e1002164 10.1371/journal.pgen.1002164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart A. L., Fishman L., Willis J. H. (2006). A simple genetic incompatibility causes hybrid male sterility in mimulus. Genetics 172 2465–2479. 10.1534/genetics.105.053686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart A. L., Flagel L. E. (2015). Evidence of natural selection acting on a polymorphic hybrid incompatibility locus in mimulus. Genetics 199 543–554. 10.1534/genetics.114.171819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Kim S. T., Chae E., Bomblies K., Zaidem M., Smith L. M., et al. (2014). Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 Alleles. PLoS Genet. 10:e1004459 10.1371/journal.pgen.1004459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar S. M. S., Singh B. (1998). Hybrid chlorosis in wheat rye crosses. Euphytica 99 1–4. 10.1023/A:1018353816039 [DOI] [Google Scholar]
- Traw M. B., Bergelson J. (2010). Plant immune system incompatibility and the distribution of enemies in natural hybrid zones. Curr. Opin. Plant Biol. 13 466–471. 10.1016/j.pbi.2010.04.009 [DOI] [PubMed] [Google Scholar]
- Uyttewaal M., Arnal N., Quadrado M., Martin-Canadell A., Vrielynck N., Hiard S., et al. (2008). Characterization of Raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for Ogura cytoplasmic male sterility. Plant Cell 20 3331–3345. 10.1105/tpc.107.057208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana P. B., Guan X. J., Ingram R. S., Tilghman S. M. (1998). Genomic imprinting is disrupted in interspecific Peromyscus hybrids. Nat. Genet. 20 362–365. 10.1038/3833 [DOI] [PubMed] [Google Scholar]
- Walia H., Josefsson C., Dilkes B., Kirkbride R., Harada J., Comai L. (2009). Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr. Biol. 19 1128–1132. 10.1016/j.cub.2009.05.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xu H., Li N., Fan F., Wang L., Zhu Y., et al. (2015). Artificial selection of Gn1a plays an important role in improving rice yields across different ecological regions. Rice 8:37 10.1186/s12284-015-0071-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H. (2011). Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl.) 10863–10870. 10.1073/pnas.1102343108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J. B., Oakey R. J., Feil R. (2014). Imprinted gene expression in hybrids: perturbed mechanisms and evolutionary implications. Heredity (Edinb) 113 167–175. 10.1038/hdy.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P., Jiang H., Wang G., Santos-González J., Köhler C. (2015). Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. Elife 4 1–14. 10.7554/eLife.10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. M., Lloyd D., Lowry D. B., Macnair M. R., Willis J. H. (2013). Indirect evolution of hybrid lethality due to linkage with selected locus in Mimulus guttatus. PLoS Biol. 11:e1001497 10.1371/journal.pbio.1001497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Kurata N., Tanoue H., Shimokawa T., Umehara Y., Yano M., et al. (1998). Physical mapping of duplicated genomic regions of two chromosome ends in rice. Genetics 150 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata Y., Yamamoto E., Aya K., Win K. T., Doi K., Sobrizal, et al. (2010). Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc. Natl. Acad. Sci. U.S.A. 107 1494–1499. 10.1073/pnas.0908283107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhao X., Cheng K., Du H., Ouyang Y., Chen J., et al. (2012). A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337 1336–1340. 10.1126/science.1223702 [DOI] [PubMed] [Google Scholar]
- Yi B., Chen Y., Lei S., Tu J., Fu T. (2006). Fine mapping of the recessive genic male-sterile gene (Bnms1) in Brassica napus L. Theor. Appl. Genet. 113 643–650. 10.1007/s00122-006-0328-9 [DOI] [PubMed] [Google Scholar]
- Yi B., Zeng F., Lei S., Chen Y., Yao X., Zhu Y., et al. (2010). Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. Plant J. 63 925–938. 10.1111/j.1365-313X.2010.04289.x [DOI] [PubMed] [Google Scholar]


