Abstract
We hypothesized that microRNA binding site single nucleotide polymorphisms (SNPs) in fibroblast growth factors (FGFs) and their receptor genes (FGFRs) may affect microRNA and mRNA interactions and are thereby associated with susceptibility of non-syndromic orofacial cleft (NSOC). Ten SNPs among the FGF and FGFR genes were selected and their associations with NSOC susceptibility were investigated in a case-control study of 602 patients with NSOC and 605 healthy controls. FGF2/rs1048201, FGF5/rs3733336 and FGF9/rs546782 showed suggestive association with NSOC susceptibility. In the combination analysis, the observed odds ratios (ORs) decreased with the number of protective alleles (rs1048201-T, rs3733336-G and rs546782-T) but were not statistically significant beyond the first comparison. Hsa-miRNA-496, hsa-miRNA-145 and hsa-miRNA-187 were predicted to be miRNAs with binding sites within/near these SNPs and were expressed in lip tissues. Decreased FGF2, FGF5 and FGF9 expression was observed in three cell lines transfected with the corresponding miRNAs. Moreover, the three SNPs could contribute to differential binding efficacy between hsa-miRNA-496 and FGF2, hsa-miRNA-145 and FGF5, hsa-miRNA-187 and FGF9 in luciferase assay. The results suggest that FGF2/rs1048201, FGF5/rs3733336 and FGF9/rs546782 are associated with the risk of NSOC and that these miRNA-FGF interactions may affect NSOC development.
Non-syndromic orofacial cleft (NSOC) is the most common facial deformity and the main type of orofacial cleft. NSOC affects approximately one in seven hundred neonates worldwide with substantial ethnic and geographic variations1. NSOC can affect not only oral functions but also the health-related quality of life. The medical expenditures for NSOC are approximately decupled compared with unaffected children and also include the costs required for caregivers, dental care, speech therapy, and special education2. NSOC etiology involves more complicated heredity and environmental factors compared with the syndromic category3.
MicroRNAs (miRNAs) are endogenous non-coding RNAs that are approximately 20–25 nt in length. miRNAs play important roles in living organisms by pairing with mRNAs to guide their post-transcriptional expression and inhibit their translation, resulting in the destabilization of their target mRNAs4. miRNAs are widely expressed in developing murine embryonic orofacial tissues, and some play developmental roles by targeting genes involved in critical orofacial development processes, such as cell proliferation, apoptosis, cell adhesion, differentiation, and epithelial-mesenchymal transition (EMT)5. For instance, miRNA-140 is a critical regulator of the Pdgf signaling pathway during palatogenesis6 and the miRNA-17-92 cluster regulates the expression pattern of critical T-box transcriptional regulators during midface development7. Therefore, the miRNA-mRNA interaction is an important mechanism in orofacial development. However, this interaction can be affected by single nucleotide polymorphisms (SNPs) within or near the binding site (commonly referred to as miRNA binding site SNPs) that may alter the thermodynamic interplay between them8. For example, a mutation 10bp away from a predicted miRNA-140 binding site in the PDGFRa 3′-UTR resulted in increased binding affinity for miRNA-140 and thus depressed PDGFRa mRNA expression during human palate development9. Similarly, our previous study showed that the differential interaction between miR-3649 and its polymorphic target in the 3′-UTR of the muscle segment homeobox1 (MSX1) played a crucial role in NSOC susceptibility10.
Fibroblast growth factors (FGFs) and fibroblast growth factor receptors (FGFRs) have critical functions in orofacial development11. Highly conserved throughout metazoan evolution, these growth factors have been detected in thousands of animal species ranging from nematodes and zebrafish to mice and humans12. There are 18 mammalian FGF genes (FGF1–FGF10and FGF16–FGF23) and four primary FGFR genes (FGFR1–FGFR4) that encode ligands and receptors respectively, which initiate the FGF signaling cascade. These genes are especially known for their roles in the induction and migration of neural crest cells, skeletogenesis and EMT, all of which are fundamental processes for craniofacial development13. Aberrations in this pathway are associated with many types of craniofacial congenital disorders, including the syndromic forms of cleft lip and palate such as Kallman syndrome, Crouzon syndrome and Apert syndrome14. Furthermore, FGF signaling is closely integrated with other craniofacial development-related pathways, such as BMP, SHH, WNT, TGF and MSX15. Therefore, FGF signaling pathway has steady functions in lip and palate morphogenesis, with perturbation of its expression patterns sometimes leading to cleft pathogenesis16. To date, several studies have found that SNPs in FGFs and FGFRs are associated with NSOC development. For example, SNPs in FGF1 were shown to be associated with a NSOC predisposition16,17. However, the majority of these studies were conducted in Caucasians with a limited sample size and none focused on miRNA binding site SNPs in FGFs and FGFRs that might also contribute considerable genetic contributions to NSOC. To address these issues, in this study we systematically selected miRNA-binding site SNPs in FGFs and FGFRs and investigated their associations with the risk of NSOC in a case-control study. Additionally, functional studies were performed to interpret the functions of the associated SNPs.
Results
Characteristics of the study subjects
The detailed characteristics of the study subjects are shown in Supplementary Table S1. There was no statistical difference in gender distribution between the cases and controls. The difference of age distributions is of no concern because oral clefts are birth defects, and the measurements of gene expressions are taken among the affected only. The case group could be divided into three subgroups according to the clinical phenotypes [cleft lip only (CLO), cleft lip with cleft palate (CLP) and cleft palate only (CPO), accounting for 39.5%, 50.6% and 9.3% of the cases, respectively].
General SNP information
As shown in Table 1, ten candidate SNPs located in seven genes (FGFR1, FGFR2, FGF2, FGF5, FGF6, FGF9 and FGF17) were identified in the study based on the inclusion criteria; their potential binding miRNAs are also listed. All of these SNPs were successfully genotyped with call rates greater than 95%. The genotype frequencies among the controls were consistent with the Hardy-Weinberg equilibrium (HWE, P > 0.05) with the exception of rs2241286 (P = 0.009) based on the preliminary analyses (Table 2).
Table 1. Primary information of selected SNPs.
| ID | rs No. | Chr | Position | Gene | Allele | MAF in CHB | Predicted miRNAsa |
|---|---|---|---|---|---|---|---|
| 1 | rs13317 | 8 | 38388671 | FGFR1 | C > T | T: 0.427 | has-miR-3128 |
| 2 | rs1476215 | 4 | 124037721 | FGF2 | T > A | A: 0.089 | hsa-miR-196a |
| 3 | rs1048201 | 4 | 124033758 | FGF2 | C > T | T: 0.375 | hsa-miR-496 |
| 4 | rs1047057 | 10 | 123229102 | FGFR2 | G > A | A: 0.375 | hsa-miR-575, hsa-miR-770-5p |
| 5 | rs4690150 | 4 | 81430594 | FGF5 | C > G | G: 0.387 | hsa-miR-452 |
| 6 | rs6838203 | 4 | 81428016 | FGF5 | T > A | A: 0.337 | hsa-miR-1243, hsa-miR-509-3p |
| 7 | rs3733336 | 4 | 81426987 | FGF5 | A > G | G: 0.253 | hsa-miR-23a |
| 8 | rs2241286 | 12 | 4413622 | FGF6 | G > A | A: 0.232 | hsa-miR-4677-3p |
| 9 | rs546782 | 13 | 21173946 | FGF9 | A > T | T: 0.083 | hsa-miR-187 |
| 10 | rs3176304 | 8 | 21961718 | FGF17 | G > C | C: 0.275 | hsa-miR-324-3p |
Chr, Chromosome; MAF, minor allele frequency; CHB, Chinese Han Beijing.
aSNPinfo Web Server ( http://manticore.niehs.nih.gov/snpinfo/snpfunc.htm) and mirSNP ( http://bioinfo.bjmu.edu.cn/mirsnp/search/).
Table 2. Association of SNPs with non-syndromic orofacial cleft susceptibility.
| SNP ID | P value for HWE | Allelic Comparison |
Co-dominant model |
Additive model |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Palla | OR, 95%CI | Phetb | OR, 95%CI | Phomc | OR, 95%CI | Paddd | OR, 95%CI | ||
| rs13317 | 0.474 | 0.935 | 0.99 [0.84, 1.18] | 0.919 | 1.01 [0.80, 1.29] | 0.859 | 0.97 [0.66, 1.42] | 0.934 | 0.99 [0.84, 1.18] |
| rs1476215 | 0.522 | 0.414 | 1.17 [0.80, 1.70] | 0.302 | 1.23 [0.83, 1.81] | 0.999 | — | 0.406 | 1.18 [0.80, 1.72] |
| rs1048201 | 1.000 | 0.026 | 0.83 [0.71, 0.98] | 0.007 | 0.70 [0.13, 0.54] | 0.049 | 0.72 [0.17, 0.52] | 0.026 | 0.83 [0.71, 0.98] |
| rs1047057 | 0.596 | 0.342 | 1.08 [0.92, 1.27] | 0.194 | 1.19 [0.92, 1.53] | 0.450 | 1.14 [0.81, 1.59] | 0.337 | 1.08 [0.92, 1.28] |
| rs4690150 | 0.253 | 0.49 | 0.94 [0.80, 1.11] | 0.785 | 0.97 [0.75, 1.25] | 0.457 | 0.88 [0.18, 0.62] | 0.482 | 0.94 [0.80, 1.11] |
| rs6838203 | 0.852 | 0.469 | 1.06 [0.90, 1.26] | 0.039 | 1.30 [1.01, 1.67] | 0.952 | 0.99 [0.69, 1.42] | 0.468 | 1.06 [0.90, 1.26] |
| rs3733336 | 0.181 | 0.001 | 0.73 [0.60, 0.88] | 0.013 | 0.74 [0.28, 0.83] | 0.008 | 0.48 [0.28, 0.83] | < 0.001 | 0.72 [0.59, 0.87] |
| rs2241286 | 0.009 | — | — | — | — | — | — | — | — |
| rs546782 | 0.509 | 0.040 | 0.57 [0.33, 0.98] | 0.066 | 0.60 [0.34, 1.04] | — | — | 0.043 | 0.57 [0.33, 0.98] |
| rs3176304 | 1.000 | 0.689 | 1.04 [0.86, 1.26] | 0.287 | 1.14 [0.90, 1.45] | 0.613 | 0.88 [0.53, 1.46] | 0.689 | 1.04 [0.86, 1.26] |
SNP, single nucleotide polymorphism; Chr, chromosome; OR, odds ratio; 95%CI, 95% confidence interval.
aP value of allelic comparison; rs1048201- T vs. C, rs3733336- G vs. A, rs546782-T vs. A.
bP value of heterozygous comparison; rs1048201 - CT vs. CC, rs3733336 - AG vs. AA, rs546782 - AT vs. AA.
cP value of homozygous comparison; rs1048201 - TT vs. CC, rs3733336 - GG vs. AA, rs546782-TT vs. AA.
dP value of additive model analysis.
Association Studies
Overall association analysis
After excluding rs2241286 from further analysis (deviation from HWE), nine SNPs remained in the association analysis with NSOC susceptibility. Allelic comparisons and genotypic comparisons including the heterozygous, homozygous and additive models were applied. Among them, three SNPs [rs1048201 on FGF2 (Padd = 0.026), rs3733336 on FGF5 (Padd < 0.001), and rs546782 on FGF9 (Padd = 0.043)] showed significant associations with NSOC susceptibility in the additive model (Table 2). For rs1048201, a significant association was observed in all genetic models with the T allele associated with a reduced NSOC risk (ORall = 0.83, 95% CI = 0.71–0.98; ORhet = 0.70, 95% CI = 0.13–0.54; ORhom = 0.72, 95% CI = 0.17–0.52; ORadd = 0.83, 95% CI = 0.71–0.98). A protective effect of rs3733336-G was also found in all of the models (ORall = 0.73, 95% CI = 0.60–0.88; ORhet = 0.74, 95% CI = 0.28–0.83; ORhom = 0.48, 95% CI = 0.28–0.83; ORadd = 0.72, 95% CI = 0.59–0.87). A notable relationship between rs546782 and the risk of NSOC was observed in the allelic comparison (ORall = 0.57, 95% CI = 0.33–0.98) and additive comparison model (ORadd = 0.57, 95% CI = 0.33–0.98).
Stratified analysis
Appreciable differences were identified in the CLO, CLP and CPO etiology, which prompted us to conduct stratified analyses18. As shown in Supplementary Table S2, 232, 289 and 49 patients were included in the CLO, CLP and CPO groups, respectively. Protective effects of the rs1048201-T allele were observed for CLO and CLP (Padd = 0.028 for CLO and Phet = 0.046 for CLP) and the rs3733336-G allele for CLO and CPO (Padd = 0.012 for CLO and Padd = 0.017 for CPO). A lack of association was observed between rs546782 and any NSOC subgroups (Supplementary Table S3). The multinomial logistic regression of three subtypes showed the similar results by additive model (Supplementary Table S4).
Combination analysis
Next, we evaluated the combined effects of the three SNPs that were significant in the overall analysis. The observed ORs decreased with the number of protective alleles (rs1048201-T, rs3733336-G and rs546782-T) but were not statistically significant beyond the first comparison. Individuals carrying one, two, and three to six protective alleles (rs1048201-T, rs3733336-G and rs546782-T) had a 0.67, 0.53 and 0.48-fold decreased risk of NSOC compared with individuals carrying no protective allele, respectively (P < 0.001 in the multi degree-of-freedom likelihood ratio test). However, there was no statistically significant difference of the NSOC risk between the groups carrying one, two, and three to six protective alleles (Table 3).
Table 3. Combined effects of three SNPs on non-syndromic orofacial cleft.
| Protective allele numbera | Case (%) | Control (%) | OR (95%CI) | P |
|---|---|---|---|---|
| 0 | 118 (20.59) | 77 (13.05) | Reference | |
| 1 | 220 (38.39) | 214 (36.27) | 0.67 (0.48–0.95) | 0.023 |
| 2 | 170 (29.67) | 210 (35.59) | 0.53 (0.37–0.75) | <0.001 |
| 3–6 | 65 (11.34) | 89 (15.08) | 0.48 (0.31–0.73) | <0.001 |
| 1–6 | 455 (79.41) | 513 (86.95) | 0.58 (0.42–0.79) | <0.001 |
| Pb < 0.001 | ||||
| 2 vs. 1 | — | — | 0.78 (0.59–1.03) | 0.083 |
| 3–6 vs. 2 | — | — | 0.91 (0.62–1.33) | 0.653 |
| 3–6 vs. 1 | — | — | 0.71 (0.49–1.04) | 0.081 |
ars1048201-T, rs3733336-G and rs546782-T were assumed as protective alleles based on the association study in Table 2.
bP value of multi degree-of-freedom likelihood ratio test.
As shown in Supplementary Table S5, combined effects were also assessed in the three NSOC subgroups. Individuals with one protective allele were associated with a decreased risk of CLP. Associations with decreased CLO, CLP and CPO risks were observed with two or more protective alleles (P = 0.048 in CLO, P = 0.001 in CLP and P = 0.538 in CPO).
Functional Studies
We subsequently conducted functional analysis on the three SNPs (rs1048201, rs3733336, and rs546782) that had suggestive associations with the NSOC risk in association studies.
Lip tissue expression of the predicted binding miRNA partners
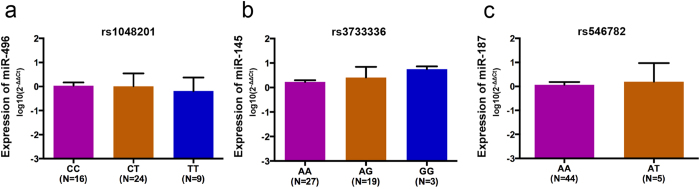
As predicted by the bioinformatics analysis and illustrated in Supplementary Figure S1, these three SNPs might affect the binding ability of hsa-miRNA-496 (miR-496), hsa-miRNA-145 (miR-145) and hsa-miRNA-187 (miR-187) to the corresponding FGFs. We collected redundant lip skin tissues from orofacial cleft plastic surgeries to confirm miRNA expression in lip tissues. As shown in Fig. 1a–c, these miRNAs were stably expressed in all lip tissues without association with the FGF2/rs1048201, FGF5/rs3733336 and FGF9/rs546782 genotypes, respectively.
Figure 1. miRNA expression in lip tissues of non-syndromic orofacial cleft cases.
(a–c) miR-496, miR-145 and miR-187 expression in 49 lip skin tissue samples from NSOC patients by qPCR, respectively. The results were normalized to U6. Error bars indicate the standard errors.
Confirmation of the interaction between the miRNAs and mRNAs in vitro
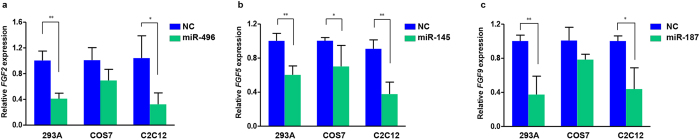
We transfected miR-496, miR-145 and miR-187 mimics into three cell lines (HEK-293A, COS7 and C2C12) to test whether the miRNAs interacted with their corresponding FGFs. As shown in Fig. 2a, FGF2 expression was significantly decreased in HEK-293A (P = 0.004) and C2C12 cell lines (P = 0.032) transfected with miR-496. Transfection of the miR-145 mimic into HEK-293A, COS7 and C2C12 cell lines resulted in significantly decreased FGF5 expression (P = 0.010, 0.020 and 0.007, respectively) (Fig. 2b). FGF9 expression was significantly decreased in HEK-293A (P = 0.009) and C2C12 cell lines (P = 0.019) transfected with miR-187 (Fig. 2c).
Figure 2. Quantitative real-time polymerase chain reaction analysis of FGF2, FGF5 and FGF9 expression in vitro.
(*P < 0.05, **P < 0.01, ***P < 0.001). (a) FGF2 expression levels in HEK-293A cells, COS7 cells and C2C12 cells transfected with the miR-496 mimics and nonsense RNA fragments (NC). (b) FGF5 expression levels in HEK-293A cells, COS7 cells and C2C12 cells transfected with the miR-145 mimics and nonsense RNA fragments (NC). (c) FGF9 expression levels in HEK-293A cells, COS7 cells and C2C12 cells transfected with the miR-187 mimics and nonsense RNA fragments (NC).
miRNA-mRNA interactions directed by SNPs
The dual luciferase reporter assay was performed to test whether these three SNPs contributed to the different binding efficiencies between the miRNAs and mRNAs.
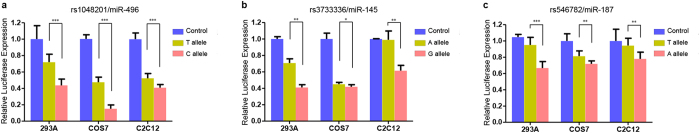
The reporter gene assay suggested that the T allele of rs1048201 attenuated miR-496 binding in HEK-293A, COS7 and C2C12 cell lines compared with the C allele (P < 0.001 in all cell lines, Fig. 3a), whereas the G allele of rs3733336 had stronger binding affinity for miR-145 than the A allele in HEK-293A (P = 0.005), COS7 (P = 0.049) and C2C12 (P = 0.009) cell lines (Fig. 3b). The rs546782-T allele showed lower binding affinity for miR-187 compared with the A allele (P < 0.001 in HEK-293A, P = 0.001 in COS7, and P = 0.001 in C2C12 cell lines, Fig. 3c).
Figure 3. Binding ability assay of plasmids constructed with the 3′-UTR fragments of FGF2, FGF5 and FGF9 and the miRNAs in HEK-293A cells, COS7 cells and C2C12 cells.
A plasmid construct with a nonsense sequence was used as the negative control (NC). The firefly luciferase to Renilla luciferase ratio was considered the relative luciferase expression. Independent triplicate experiments were performed. (*P < 0.05, **P < 0.01, ***P < 0.001). (a) The plasmid containing the C/T allele of rs1048201 was co-transfected with miR-496. (b) The plasmid containing the A/G allele of rs3733336 was co-transfected with miR-145. (c) The plasmid containing the A/T allele of rs546782 was co-transfected with miR-187.
Discussion
miRNA binding site SNPs can affect miRNA interactions with their putative target sites due to allele differences. This type of influence may be functional and alter the expression level of the target genes, thereby contributing to altered phenotypes19. In the present study, ten miRNA-binding site SNPs in seven FGF/FGFR genes were selected and successfully genotyped in a case-control study. One of the SNPs (rs2241286) was excluded from the analysis due to its deviation from the HWE. Of the remaining nine SNPs, three SNPs (FGF2/rs1048201, FGF5/rs3733336, and FGF9/rs546782) exhibited evidence of association with NSOC in the Chinese Han population. Additionally, FGF2/rs1048201 and FGF5/rs3733336 could be favorable indicators of an individual’s susceptibility to specific subgroups. The combined analysis showed that the protective effects against NSOC strengthened with the increasing number of protective alleles.
The protective T allele of rs1048201 decreased the FGF2 binding capacity for miR-496 in vitro compared with the C allele, resulting in a higher mRNA level. These results corroborated the increased expression of FGF2 during normal cranial fusion20. FGF2 was demonstrated to have profitable effects on cell proliferation and angiogenesis in periodontal tissues and oral mucosa21 as well as cranial suture fusion22. Mutations contributing to impaired transcription in the non-coding region of FGF2 were associated with the development of orofacial clefts23. Furthermore, FGF2 treatment promoted rat osteoblast attachment and produced enhanced suture fusion in calvarial organ culture24.
FGF5 has been recognized as a development-related gene. It is widely expressed in embryos and participates in the interaction between the epithelium and mesenchymal cells, the formation of the neural crest, angiogenesis and cell proliferation13,25. In the present study, rs3733336 in the FGF5 3′-UTR was identified as a functional SNP associated with the risk of NSOC. The G allele was recognized as a protective allele in the association study. The initially predicted hsa-miRNA-23a (miR-23a) (Table 1) was not demonstrated to bind FGF5 as predicted (Supplementary Figure S2). We found that there was a binding site for hsa-miRNA-145 (miR-145) at 30bp upstream of rs3733336 in further analysis by Targetscan ( http://www.targetscan.org) and SNPinfo Web Server ( http://manticore.niehs.nih.gov/snpinfo/snpfunc.htm). We hypothesized that rs3733336 may affect interaction between miR-145 and FGF5. Finally, the luciferase reporter assay showed that the transcriptional activity of the reporter gene with the G allele was significantly reduced compared with the A allele in all three cell lines, indicating that the mutant allele in the 3′-UTR activated FGF5 degradation as a protection mechanism. Thus, the interaction between rs3733336-G and miR-145 acted as a possible profitable mechanism to reduce the risk of NSOC.
FGF9 regulated cell proliferation in the palatal mesenchyme during mouse palatogenesis and exhibited an extremely high expression level in the epithelial-mesenchymal interaction phase of the mouse embryo palate processes26. Similar to FGF2, an increased FGF9 level is profitable. For instance, a total of 40% of Fgf9−/− mouse embryos exhibited cleft palates in the study of Colvin et al.27. SNPs in the FGF9 3′-UTR were identified as critical markers for gene-gene interactions in NSOC etiology16. However, the biological mechanism has not been clarified to date. In the present study, the T allele of rs546782 broke down the pairing with miR-187 and exhibited a protective effect against NSOC.
The experimental design and potential mechanism underlying the actions by which these FGF and FGFR SNPs contribute to NSOC is illustrated in Supplementary Figure S3. Taken together, our findings demonstrate that FGF2/rs1048201, FGF5/rs3733336 and FGF9/rs546782 individually and jointly contribute to the risk of NSOC and the NSOC subgroups. These SNPs affected the binding ability of miRNAs to their target genes and then modified the expression levels of the FGF genes to contribute to NSOC susceptibility. These studies help illustrate the mechanism underlying NSOC. However, two major limitations should be addressed. First, we did not perform multiple corrections in the present study, which might contribute to false positive results. Additional replication studies are needed to verify our findings. Second, further investigations between these three SNPs and the risk of CPO are warranted due to the limited CPO sample size in the current study. Therefore, extensive studies are warranted to obtain a greater understanding of our findings.
Material and Methods
Sample recruitment
This study is an ongoing study approved by the Ethics Review Committee of Nanjing Medical University (NJMUERC [2008] No. 20). All the methods were carried out in accordance with the protocols approved by the ethics committee. All subjects in both groups voluntarily joined this study and provided informed consent. The present study consisted of 602 NSOC cases and 605 gender-matched healthy controls recruited from three affiliated hospitals of Nanjing Medical University (the Stomatological Hospital of Jiangsu Province, Xuzhou First People’s Hospital, and Nanjing Children’s Hospital) between August 2008 and August 201228. All patients were clinically assessed according to the detailed diagnostic information obtained from the medical records and physical examinations. Only patients who had an isolated oral cleft without syndromic symptoms or other major congenital defects were included. The controls were examined by two experienced oral surgeons to ensure that they had no cleft, hypodontia or other congenital anomalies. Approximately 3ml of venous blood was drawn from each participant for the genotyping analysis.
SNP selection
Common SNPs (minor allele frequency, MAF ≥ 5%) of 18 FGFs (FGF1–FGF10 and FGF16–FGF23) and 4 FGFRs (FGFR1-FGFR4) of Homo sapiens in Chinese Han population based on the dbSNP database ( http://www.ncbi.nlm.nih.gov/projects/SNP/index.html) and HapMap Project database ( http://hapmap.ncbi.nlm.nih.gov/) were screened from gene regions (including ± 2kb). Then, in silico bioinformatics predictions from the SNPinfo Web Server ( http://manticore.niehs.nih.gov/snpinfo/snpfunc.htm) and mirSNP ( http://bioinfo.bjmu.edu.cn/mirsnp/search/) were collectively applied to select the miRNA binding site SNPs. Linkage disequilibrium (LD) analysis with an r2 threshold of 0.80 was applied to filter these SNPs. Finally, ten SNPs were analyzed in our study. The characteristic information of the final identified SNPs as well as their corresponding potential binding miRNAs are listed in Table 1.
DNA extraction and genotyping
The conventional phenol-chloroform method was conducted to extract genomic DNA as previously described10,28. All samples with combinations of two negative and two positive references were genotyped using a blind method based on double ligation and multiplex fluorescence polymerase chain reaction (PCR) with a custom-by-design 2 × 48-Plex SNPscanTM Kit with the support of Genesky Biotechnologies Inc. (Shanghai, China)29. A total of 5% of the samples were randomly repeated for quality control, and the results were totally concordant.
Quantitative PCR of predicted miRNAs in lip skin tissues
A total of 49 redundant lip skin tissues of cleft cases were obtained from plastic surgeries and frozen in liquid nitrogen. Total RNA was extracted by the conventional TRIzol method (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The miRNA expression levels were measured by quantitative PCR (qPCR) with the SYBR Green Real-Time PCR Master Mix kit (Takara, Shiga, Japan) on an ABI 7900 System (Applied Biosystems, Foster City, CA, USA) using the transcription level of U6 as the internal control30 (Primers in Supplementary Table S6). The relative gene expression was quantified by the comparative Ct method also referred to as the 2−ΔΔCt method31. All PCR assays were performed in triplicate to verify the results.
Cell culture
HEK-293A, COS7 and C2C12 cell lines were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China), and cultured in Dulbecco's modified Eagle’s medium (Gibco, Foster City, CA, USA) with 10% fetal bovine serum (Gibco).
miRNA transfection and qPCR
Forty-eight hours after transfection with the miRNA mimics using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as recommended by the manufacturer, the cells were harvested for qPCR analysis.
Total RNA was extracted from the cells using the conventional TRIzol method (Invitrogen) according to the manufacturer’s instruction. The FGF2, FGF5 and FGF9 expression levels were measured by qPCR with the SYBR Green Real-Time PCR Master Mix kit (Takara, Shiga, Japan) on ABI 7900 System (Applied Biosystems). The GAPDH transcription level was detected as the internal control (Primers in Supplementary Table S6). All PCR assays were performed in triplicate to verify the results.
Luciferase reporter plasmid construction
The FGF2, FGF5 and FGF9 3′-UTR fragments containing the major alleles of SNPs were inserted downstream of the luciferase gene between two restrictive sites in the luciferase reporter psiCHECKTM-2 vector (Promega, Madison, WI, USA). The minor alleles were generated by the site-specific mutagenesis method using constructs containing the major alleles as the template. The accuracy of the recombinant plasmids was verified by DNA sequencing (the sequencing results of the fragments are shown in Supplementary Figure S4, ABC). The psiCHECKTM-2 vector with a random inserted fragment was applied as the negative control.
Transient co-transfection and dual-luciferase reporter assay
Plasmids containing the wild type or mutant type allele were co-transfected with the miRNA mimics using Lipofectamine 2000 (Invitrogen). The luciferase activity in the lysates was quantified forty-eight hours after transfection with a dual-luciferase reporter assay system (Promega). The luminescent reaction of the Renilla luciferase was simultaneously activated after the firefly luciferase reporter was measured as a stabilized luminescent signal. The firefly luciferase to Renilla luciferase ratio was considered the relative reporter activity. Independent triplicate experiments were performed for each plasmid construct.
Statistical analysis
Pairwise linkage disequilibrium (LD) was computed as r2 for all SNPs by the Haploview program. Student’s t test was employed for continuous values. The gender distribution between cases and controls was evaluated with a two-sided Chi-square test, and the HWE among the controls was calculated using a goodness-of-fit Chi-square test. All results were two-sided. P < 0.05 was considered statistically significant. All tests were performed with version 9.1 of the SAS® software (SAS Institute Inc., Cary, NC, USA).
The association between the SNPs and the risk of NSOC and subgroups was measured by the odds ratio (OR) and 95% confidence interval (95% CI)32,33. Four kinds of genetic models were adopted. In Table 2, if T is the variant of interest (the mutant allele), and C is the dominant allele, a 2 by 2 table and an unconditional logistic regression with one degree of freedom could be used to determine statistical significance of allele frequencies under the assumption of an allelic comparison33. A heterozygous comparison presented the comparison of CT genotypes with CC genotypes. Likewise, comparison of CC with TT genotypes was assumed as homozygous comparison. Scores of 0, 1, and 2 assigned to genotype CC, CT, and TT respectively and ORs calculated by unconditional logistic regression model was a test for association between the variant allele and the disease named as additive model.
Additional Information
How to cite this article: Li, D. et al. Associations between microRNA binding site SNPs in FGFs and FGFRs and the risk of non-syndromic orofacial cleft. Sci. Rep. 6, 31054; doi: 10.1038/srep31054 (2016).
Supplementary Material
Acknowledgments
The study was supported by the following fundings: National Natural Science Foundation of China (81570959, 81170981, 81230022, 81400546 and 81200808), Ph.D. Programs Foundation of Ministry of Education of China ( 20123234120004, 20113234110003), China Postdoctoral Science Foundation (201104537, 20100481164), and the Priority Academic Program Development of JiangSu Higher Education Institutions (PAPD2014-37), Xuzhou science and technology project (XZZD1245), The Natural Science Foundation of JiangSu Province (BK2012447 and 15KJA320002), The open project foundation of Jiangsu Key Laboratory of Oral Diseases(JSKLOD-KF-1504), Qinglan Project for Yongchu Pan (2016-2019). The authors thank Professor Melin Wang and Juncheng Dai (School of Public Health, Nanjing Medical University, Nanjing, China) for consultation regarding the statistical analysis and are grateful to all of the participants, research staff and students who contributed to this study.
Footnotes
Author Contributions D.L, Y.H. and M.X. conducted the experiments and prepared the figures. D.L. wrote the main manuscript. H.Z. and L.M. made the arrangement of the work and analyzed the results. Z.W, H.J. and W.Z recruited the study subjects. L.W. and Y.P. searched the related papers, conceived the experiments and revised the manuscript. All authors reviewed the manuscript.
References
- Sun Y. et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nature communications 6, 6414, doi: 10.1038/ncomms7414 (2015). [DOI] [PubMed] [Google Scholar]
- Wehby G. L. & Cassell C. H. The impact of orofacial clefts on quality of life and healthcare use and costs. Oral diseases 16, 3–10, doi: 10.1111/j.1601-0825.2009.01588.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey P. A., Little J., Munger R. G., Dixon M. J. & Shaw W. C. Cleft lip and palate. Lancet 374, 1773–1785, doi: 10.1016/S0140-6736(09)60695-4 (2009). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233, doi: 10.1016/j.cell.2009.01.002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P. et al. Developmental microRNA expression profiling of murine embryonic orofacial tissue. Birth defects research. Part A, Clinical and molecular teratology 88, 511–534, doi: 10.1002/bdra.20684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart J. K. et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nature genetics 40, 290–298, doi: 10.1038/ng.82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. MicroRNA-17-92, a direct Ap-2alpha transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS genetics 9, e1003785, doi: 10.1371/journal.pgen.1003785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan B. M., Robles A. I. & Harris C. C. Genetic variation in microRNA networks: the implications for cancer research. Nature reviews. Cancer 10, 389–402, doi: 10.1038/nrc2867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattanasopha S. et al. PDGFRa mutations in humans with isolated cleft palate. European journal of human genetics: EJHG 20, 1058–1062, doi: 10.1038/ejhg.2012.55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. et al. A miRNA-binding-site SNP of MSX1 is Associated with NSOC Susceptibility. Journal of dental research 93, 559–564, doi: 10.1177/0022034514527617 (2014). [DOI] [PubMed] [Google Scholar]
- Teven C. M., Farina E. M., Rivas J. & Reid R. R. Fibroblast growth factor (FGF) signaling in development and skeletal diseases. Genes & Diseases 1, 199–213, doi: 10.1016/j.gendis.2014.09.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R. & Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nature reviews. Molecular cell biology 14, 166–180, doi: 10.1038/nrm3528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauws E. & Stanier P. FGF signalling and SUMO modification: new players in the aetiology of cleft lip and/or palate. Trends in genetics: TIG 23, 631–640, doi: 10.1016/j.tig.2007.09.002 (2007). [DOI] [PubMed] [Google Scholar]
- Francois-Fiquet C. et al. Role of angiogenesis-related genes in cleft lip/palate: review of the literature. International journal of pediatric otorhinolaryngology 78, 1579–1585, doi: 10.1016/j.ijporl.2014.08.001 (2014). [DOI] [PubMed] [Google Scholar]
- Li Z., Yu M. & Tian W. An inductive signalling network regulates mammalian tooth morphogenesis with implications for tooth regeneration. Cell proliferation 46, 501–508, doi: 10.1111/cpr.12051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. The FGF and FGFR Gene Family and Risk of Cleft Lip With or Without Cleft Palate. The Cleft palate-craniofacial journal: official publication of the American Cleft Palate-Craniofacial Association 50, 96–103, doi: 10.1597/11-132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqdoost Z. et al. Investigation of FGF1 and FGFR gene polymorphisms in a group of Iranian patients with nonsyndromic cleft lip with or without cleft palate. International journal of pediatric otorhinolaryngology 78, 731–736, doi: 10.1016/j.ijporl.2014.01.024 (2014). [DOI] [PubMed] [Google Scholar]
- Dixon M. J., Marazita M. L., Beaty T. H. & Murray J. C. Cleft lip and palate: understanding genetic and environmental influences. Nature reviews. Genetics 12, 167–178, doi: 10.1038/nrg2933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M. A., Liang H. & Li W. H. Human polymorphism at microRNAs and microRNA target sites. Proceedings of the National Academy of Sciences of the United States of America 104, 3300–3305, doi: 10.1073/pnas.0611347104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrara B. J., Mackool R. J., McCarthy J. G., Gittes G. K. & Longaker M. T. Immunolocalization of basic fibroblast growth factor and fibroblast growth factor receptor-1 and receptor-2 in rat cranial sutures. Plastic and reconstructive surgery 102, 1805–1817; discussion 1818-1820 (1998). [DOI] [PubMed] [Google Scholar]
- Bizenjima T. et al. Fibroblast growth factor-2 promotes healing of surgically created periodontal defects in streptozotocin-induced early diabetic rats via increasing cell proliferation and regulating angiogenesis. Journal of clinical periodontology, doi: 10.1111/jcpe.12324 (2014). [DOI] [PubMed] [Google Scholar]
- Pan A., Chang L., Nguyen A. & James A. W. A review of hedgehog signaling in cranial bone development. Frontiers in physiology 4, 61, doi: 10.3389/fphys.2013.00061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B. M. & Murray J. C. Sequence evaluation of FGF and FGFR gene conserved non-coding elements in non-syndromic cleft lip and palate cases. American journal of medical genetics. Part A 143A, 3228–3234, doi: 10.1002/ajmg.a.31965 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Ge S., Shim Y. H., Zhang C. & Cao D. Bone morphogenetic protein is required for fibroblast growth factor 2-dependent later-stage osteoblastic differentiation in cranial suture cells. International journal of clinical and experimental pathology 8, 2946–2954 (2015). [PMC free article] [PubMed] [Google Scholar]
- Haub O. & Goldfarb M. Expression of the fibroblast growth factor-5 gene in the mouse embryo. Development 112, 397–406 (1991). [DOI] [PubMed] [Google Scholar]
- Iwata J. et al. Fibroblast growth factor 9 (FGF9)-pituitary homeobox 2 (PITX2) pathway mediates transforming growth factor beta (TGFbeta) signaling to regulate cell proliferation in palatal mesenchyme during mouse palatogenesis. The Journal of biological chemistry 287, 2353–2363, doi: 10.1074/jbc.M111.280974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin J. S., White A. C., Pratt S. J. & Ornitz D. M. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development 128, 2095–2106 (2001). [DOI] [PubMed] [Google Scholar]
- Han Y. et al. The axis inhibition protein 2 polymorphisms and non-syndromic orofacial clefts susceptibility in a Chinese Han population. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology 43, 554–560, doi: 10.1111/jop.12162 (2014). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. Journal of thrombosis and haemostasis: JTH 10, 1508–1514, doi: 10.1111/j.1538-7836.2012.04815.x (2012). [DOI] [PubMed] [Google Scholar]
- Xu H. et al. Predictive Value of Serum miR-10b, miR-29c, and miR-205 as Promising Biomarkers in Esophageal Squamous Cell Carcinoma Screening. Medicine 94, e1558, doi: 10.1097/MD.0000000000001558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D. & Livak K. J. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- Balding D. J., Bishop M. & Cannings C. Handbook of Statistical Genetics, Third Edition, 1245–1250 (2008). [Google Scholar]
- Clarke G. M. et al. Basic statistical analysis in genetic case-control studies. Nat Protoc 6, 121–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.