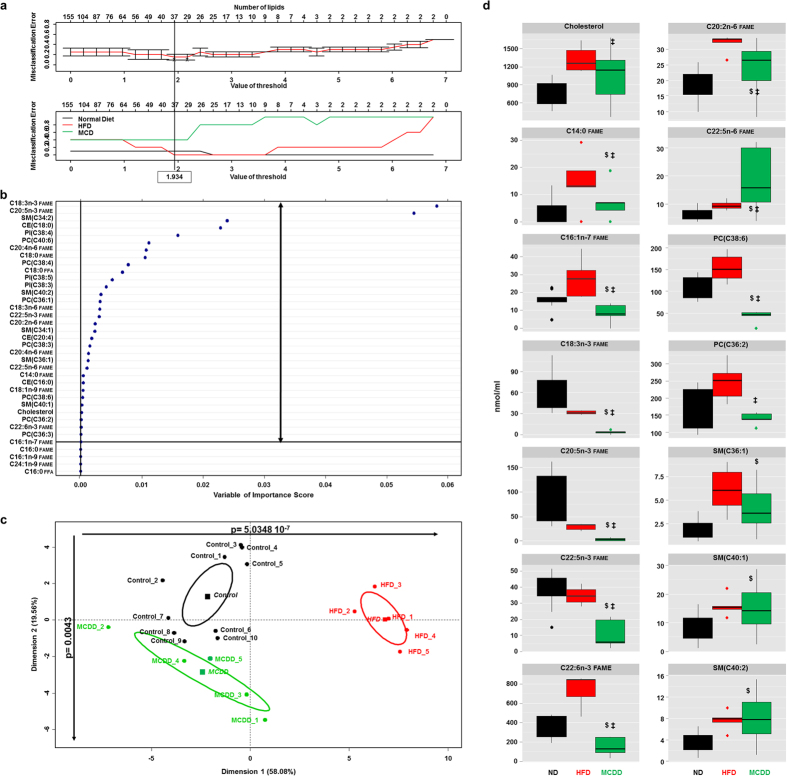
Figure 3. Specific serum lipids discriminating mice fed a MCDD.
(a) Identification of 37 lipids among 155 lipids based of the calculating threshold 1.934 to have the minimum misclassification rate between the 3 groups of mice using prediction analysis of microarray. (b) 33 lipids identified among 37 lipids discriminating the three groups of mice based on random forest analysis. (c) Principal component analysis (PCA) using 33 lipids identified in (A). Dots represent each mouse, lines are the ellipses centred to the mean (coloured squares) representing 95% interval confidence, and p the probability associated with the F- test of the analysis of variance along the axes of the first and the second dimensions (α = 0.05). (d) Boxplots of the 14 lipids from PCA discriminating mice fed MCDD. $p < 0.05 compared to mice fed ND; ‡p < 0.05 compared to mice fed HFD based on ANOVA test follow by Student t-test. CE: cholesteryl ester; HFD: high-fat diet (red); FAME: fatty acyl methyl ester; FFA: free fatty acid; MCDD: methionine-choline deficient diet (green); ND: normal diet (black); PC: phosphatidylcholine; PI: phosphatidylinositol; SM: sphingomyelin. Most representative phospholipids: PC(C38:6) = PC(18:0/20:6); PC(C36:2) = PC(18:1/18:1); SM(C36:1) = SM(18:1/18:0); SM(C40:1): SM(18:1/22:0); SM(C40:2): SM(18:1/22:1).