Highlights
-
•
Heterologous sfCherry protein was expressed in N. salina for the first time.
-
•
N. salina was transformed by particle bombardment.
-
•
Integration site of the transgene on the genome was determined by RESDA PCR.
-
•
Expression of sfCherry was confirmed by a western blotting and confocal microscopy.
Abbreviations: RESDA PCR, restriction enzyme site-directed amplification polymerase chain reaction; TAIL PCR, thermal asymmetric interlaced PCR; MARs, matrix attachment regions; EPA, eicosapentaenoic acid
Keywords: Heterologous protein expression, Particle bombardment, Restriction enzyme site-directed amplification (RESDA) PCR, sfCherry fluorescent protein
Abstract
Oleaginous microalgae of the Nannochloropsis genus are considered excellent candidates for biofuels and value-added products owing to their high biomass productivity and lipid content. Here, we report the first overexpression and detection of a heterologous sfCherry fluorescent protein in Nannochloropsis salina in order to develop a transformation toolbox for future genetic improvements. Particle bombardment was employed for transformation, and expression of Shble under the control of TUB and UEP promoters, cloned from N. salina, was used to confer resistance to Zeocin antibiotics, resulting in 5.9 and 4.7 transformants per 108 cells, respectively. Stable integration of the markers into the genome was confirmed using a restriction enzyme site-directed amplification (RESDA) PCR. The expression of sfCherry fluorescent protein was confirmed by Western blot analysis and confocal microscopy. These results suggest new possibilities of efficient genetic engineering of Nannochloropsis for the production of biofuels and other biochemicals.
1. Introduction
Growing concerns about potential energy crises and environmental problems caused by a dependence on traditional fossil fuels has increased our interest in alternative energy sources. Microalgae are among the most promising feedstocks for biofuels and natural products owing to their high lipid content and fast growth rate compared with other biofuels feedstocks, such as crops and other land plants. Moreover, they do not require arable land or fresh water for cultivation, and can grow using CO2 in flue gas, thereby reducing levels of this important greenhouse gas [1], [2].
Nannochloropsis sp. are attractive industrial production strains for biofuels and value-added products such as eicosapentaenoic acid (EPA) by virtue of their fast growth and high lipid content [3]. Various cultivation methods have been developed as part of an effort to efficiently produce biofuels from Nannochloropsis [4]. In addition, genomic and transcriptomic analyses of several Nannochloropsis strains have been reported [3]. These data can be used for genetic improvements of Nannochloropsis designed to achieve commercial production of biofuels. Recent reports have demonstrated the transformation of Nannochloropsis, indicating that the creation of ‘smart’ microalgae with high lipid productivity is now conceivable [5], [6]. Subsequent studies have reported improved transformation efficiency through delivery of a PCR product instead of an intact plasmid [7] and by using top agar for recovery [6] or conditioned medium [8]. It also has been reported that metabolic pathway related genes are knocked out using the homologous recombination method [9].
Three main types of transformation methods have generally been used for microalgae: agitation with glass beads, electroporation, and particle bombardment. Agitation with glass beads and electroporation are commonly used to transform microalgae that lack or have weakened cell walls. For microalgae with hard cell walls, such as diatoms, particle bombardment has been employed as a transformation method [10]. It is also the method of choice for organellar transformation. Notably, chloroplast transformation can be used in a microalgal cell-factory setting for producing heterologous protein and value-added products [11], [12].
An established method for localization of the transgene integration site within the host genome is thermal asymmetric interlaced polymerase chain reaction (TAIL PCR). In TAIL PCR, the insertion site can be found efficiently using nested specific primers and relatively short, arbitrary, degenerate primers [13]. RESDA (restriction enzyme site-directed) PCR is an improved version of TAIL PCR that uses degenerate primers containing restriction enzyme sites to increase the efficiency of TAIL PCR [14].
In addition, the applications of fluorescent proteins involve using them for selection marker, checking the expression level of fusion protein, and exploiting for imaging of the localization and dynamics of specific organelles [15]. However, it should be noted that proper choice of fluorescent protein is important not only for preventing the interference of autofluorescent signal from cells, but also for greater brightness and photostability [16]. Fluorescent protein has been genetically modified in order to allow them to do efficient folding and to maintain strong fluorescence [17].
In this study, we report the first overexpression and detection of a heterologous sfCherry fluorescent protein, a genetically modified mCherry, as part of an effort to develop a genetic-manipulation toolbox in Nannochloropsis salina [17]. To introduce a heterologous protein, we used particle bombardment to transform N. salina with plasmids containing a marker and/or a reporter. Transformants were further analyzed by RESDA PCR to determine if the introduced constructs were integrated into the genome [14]. Finally, in order to validate that our transformation technique resulted in the successful production of a heterologous protein, we expressed a construct of the sfCherry fluorescent protein in N. salina, and confirmed its expression by Western blot analysis and confocal fluorescence microscopy. It is expected that this work will facilitate the genetic engineering of Nannochloropsis for the production of biofuels and bioproducts.
2. Materials and methods
2.1. Microalgae strain and culture conditions
N. salina CCMP1776 (National Center for Marine Algae and Microbiota) was maintained in sterile modified F2N media [9] composed of the following: 15 g/L sea salt (Sigma–Aldrich, USA), 10 mM Tris–HCl (pH 7.6), 427.5 mg/L NaNO3, 30 mg/L NaH2PO4∙2H2O, 5 mL/L trace metal mixture (4.36 g/L Na2 EDTA∙2H2O, 3.15 g/L FeCl3∙6H2O, 10 mg/L CoCl2∙6H2O, 22 mg/L ZnSO4∙7H2O, 180 mg/L MnCl2∙4H2O, 9.8 mg/L CuSO4∙5H2O, 6.3 mg/L Na2MoO4∙2H2O), and 2.5 mL/L vitamin stock (1 mg/L vitamin B12, 1 mg/L Biotin, 200 mg/L thiamine∙HCl) [18]. Cells were cultivated in a 200 mL working volume in 250 mL Erlenmeyer baffled flasks at 25 °C with agitation (120 rpm) under fluorescent light (120 μmol photons/m2/s). Air mixed with 2% CO2 was directly supplied to the culture at a rate of 0.5 vvm (volume gas per volume medium per minute).
2.2. Vector construction
The plasmid, pNsTShble (Fig. 1a), harboring the endogenous TUB promoter, the Shble gene, which confers resistance to Zeocin (Invitrogen, USA), and the TUB terminator was constructed and used for transformation. Other plasmids used in this study include pNsUShble (UEP promoter, Shble, and UEP terminator) and pNssfCherry (TUB promoter, the gene encoding sfCherry fluorescent protein, and TUB terminator). The pNssfCherry vector also harbors the Shble gene as a selection marker flanked by the UEP promoter and the UEP terminator (Fig. 2a). All plasmids were constructed using the Gibson assembly technique [19].
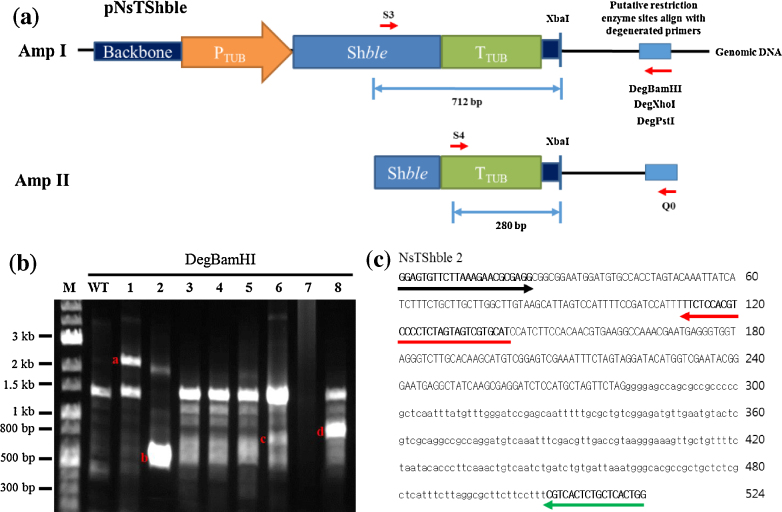
Fig. 1.
Results of RESDA PCR for N. salina wild type and NsTShble transformants 1 through 8. (a) A schematic depiction of RESDA PCR for the NsTShble transformants of N. salina. (b) The results of Amp II using DegBamHI. The bands ‘a’ to ‘d’ in the gel from DegBamHI samples were recovered and sequenced. (c) Examples of sequences obtained from NsTShble 2. Capital letters represent the plasmid region, and lowercase letters show the flanking genomic sequence. Black, red, and green arrows represent the S4, S5, and Q0 primers, respectively. Abbreviations: M, marker; WT, wild type.
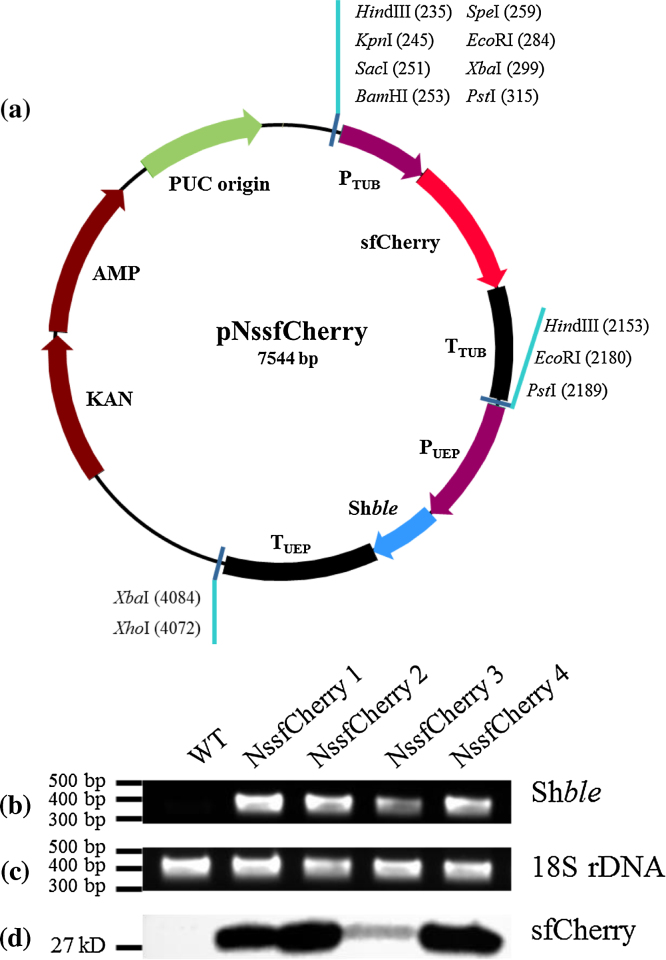
Fig. 2.
Analyses of NssfCherry transformants. (a) The vector map of pNssfCherry. Agarose gel electrophoresis for the verification of (b) the Shble PCR product and (c) 18S rDNA. (d) sfCherry fluorescent protein accumulation determined by western blotting.
2.3. Particle bombardment
N. salina cultivated in modified F2N media was harvested after 7 days in the mid-exponential phase (OD680 nm = 6), and cell number was determined using a hemocytometer. A 47-mm-diameter cellulose acetate membrane filter (Sartorius Stedim Biotech, Germany) was placed on F2N agar media, and 108 cells were placed on the membrane filter for a single bombardment. Plasmids, linearized by treatment with XbaI, were coated onto microcarrier gold particles (Bio-Rad, USA) for transformation. A mixture of plasmid, 2.5 M CaCl2, and 0.1 M spermidine was prepared in 25% glycerol containing gold particles and vigorously mixed for 3 min, after which the coated particles were allowed to settle by gravity at room temperature for 10 min. After discarding the supernatant, the coated particles were washed with 70% ethanol and then resuspended in 100% ethanol. Particle bombardment was performed using a low-pressure gene delivery system (GDS-80; Wealtec, USA) under the following conditions: 625 μg gold particles coated with 1 μg of linearized plasmid per shot by 700 psi of helium and 3 cm target distance [20]. After transformation, the cells were recovered in modified F2N media and incubated at 25 °C under fluorescent light (5 μmol photons/m2/s) without agitation for 1 day. All cells were plated onto selective F2N agar media containing 2.5 μg/mL Zeocin. After 3–4 weeks, colonies that appeared on the selective media were selected for further analyses.
2.4. Analytical procedures
2.4.1. PCR analysis of N. salina transformants
Zeocin-resistant N. salina colonies were harvested and washed with distilled water, and crude DNA was isolated using Instagene Matrix (Bio-Rad, USA) according to the manufacturer’s instructions. Briefly, 200 μL of Instagene Matrix was added to the cells and mixed. The mixtures were first incubated at 56 °C for 20 min and then at 100 °C for 8 min. After centrifugation, the supernatant was used for PCR. S1 (forward) and S2 (reverse) primers were used to detect the Shble gene in N. salina transformants (Table 1). 18S rDNA, used as a positive control, was detected using SR6 (forward) and SR9 (reverse) primers (Table 1) [21]. PCR amplification was carried out using Ex-taq polymerase (Takara, Japan) with 30 cycles of 95 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min. The expected sizes of Shble and 18S rDNA PCR fragments were 357 and 380 bp, respectively.
Table 1.
Primers used in this study.
| Primer | Sequence 5′–3′ | Purpose |
|---|---|---|
| SR6 | GTCAGAGGTGAAATTCTTGG | 18S rDNA |
| SR9 | AACTAAGAACGGCATGCAC | |
| S1 | AAGTTGACCAGTGCCGTTCCGGTG | Shble |
| S2 | CTCGGCCACGAAGTGCACGCAGTT | |
| S3 | ATGACCGAGATCGGCGAGCA | RESDA PCR |
| S4 | GGAGTGTTCTTAAAGAACGCGAGG | |
| S5 | ATGCACGACTACTAGAGGGGACGTGGAGAA | |
| DegPstI | CCAGTGAGCAGAGTGACG IIIIINNS CTGCAG W | |
| DegXhoI | CCAGTGAGCAGAGTGACG IIIIINNS CTCGAG W | |
| DegBamHI | CCAGTGAGCAGAGTGACG IIIIINNS CCTAGG W | |
| Q0 | CCAGTGAGCAGAGTGACG |
2.4.2. RESDA PCR
RESDA PCR was used to identify the insertion sites of the plasmid in the genomic DNA as described previously [14] and depicted schematically in Fig. 1a. Each PCR reaction was carried out in a final volume of 50 μL using Ex-taq polymerase (Takara, Japan). REDSA PCR consists of three stages: amplification I (Amp I), amplification II (Amp II), and re-amplification (Re-Amp). For Amp I, PCR was performed with the S3 primer and three degenerate primers (Table 1) using genomic DNA as the template. The PCR conditions for Amp I were 5 min at 96 °C followed by 20 cycles of 1 min at 95 °C, 1 min at 60 °C and 3 min at 72 °C, then 10 cycles of 1 min at 95 °C, 1 min at 40 °C, 3 min at 72 °C, and a final step of 10 min at 72 °C. For the Amp II step, PCR was conducted with S4 and Q10 primers using 1 μL of the PCR product from the Amp I step as the template. The PCR conditions for Amp II were 5 min at 96 °C followed by 35 cycles of 1 min at 95 °C, 1 min at 60 °C, 3 min at 72 °C, and a final step of 10 min at 72 °C. Insertion sites in genomic DNA were identified by sequencing the specific PCR bands between approximately 300 bp and 2 kb after purification with a gel extraction kit (Qiagen, USA). A Re-Amp step was performed to confirm that the PCR bands selected from the Amp II step were correct. The template for Re-Amp PCR was obtained from the PCR product in the Amp II analytical agarose gel by directly inserting a 200-μL pipette tip into the corresponding band and shaking the tip in 50 μL of distilled water. Re-Amp PCR was performed with S4 and S5 primers using the water containing the extracted Amp II PCR product as a template. The PCR conditions for the Re-Amp step were 5 min at 96 °C followed by 35 cycles of 30 s at 95 °C, 30 s at 60 °C, 3 min at 72 °C, and a final step of 10 min at 72 °C.
2.4.3. Western blot analysis
The expression of sfCherry in transformed cells was examined by extracting proteins with 1.5× Laemmli sample buffer, which is composed of the following components: 62.5 mM Tris–HCl (pH 7.6), 7% sodium dodecyl sulfate (SDS), 25% glycerol 5% β-mercaptoethanol, and 0.02% bromophenol blue. Cells obtained at mid-exponential phase were washed with distilled water, suspended in 1.5× Laemmli sample buffer, and then heated at 100 °C for 5 min. After centrifugation at 13,000 rpm for 5 min, the supernatants were separated by SDS-PAGE (polyacrylamide gel electrophoresis) on 4–15% gradient gels and transferred to a PVDF (polyvinylidene difluoride) membrane using a Trans-Blot Turbo system (Bio-Rad). After blocking with 5% skim milk in phosphate buffered saline (PBS), the membranes were probed with a rabbit anti-mCherry antibody (Abcam, UK). Membranes were then washed three times in 5% skim milk and then incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (Cell Signaling Technology, USA). After washing membranes three times with PBS, immunoreactive proteins were detected using enhanced chemiluminescence (ECL) reagents and the ChemiDoc system (Bio-Rad).
2.4.4. Visualization of sfCherry fluorescence
Fluorescence was visualized using an A1 Plus confocal microscope (Nikon, USA). sfCherry fluorescence of the NssfCherry transformant was detected using an mCherry filter (excitation, 561 nm; emission, 595/50 nm), and N. salina auto-fluorescence was detected using a FITC (fluorescein isothiocyanate) filter (excitation, 488 nm; emission, 525/50 nm).
3. Results and discussion
3.1. Transformation by particle bombardment and confirmation of stable integration
One major problem associated with microalgal transformation is that transgene expression is lost over time due to transgene silencing [22]. This phenomenon has hampered the development of microalgal genetic engineering techniques. The long-term stability of transformed microalgal cells without selection is one of the main issues in microalgal transformation. Compared to glass bead agitation and electroporation, bombardment appears to provide more stable transformation in the absence of selection [10]. Unfortunately, the exact mechanism how the gene delivery method affects the stability of the heterologous expression is unknown. However, there are some reports describing the long-term stability of transformants according to transformation methods. Based on statistical analysis, the long term stability of phenotype has been reported in 90.6% of papers whose authors used the bombardment method, and in 66.6% and 33.3% of those that used glass beads and electroporation, respectively [10]. This statistical analysis implies that after bombardment, the presence of exogenous DNA in the genome of the transformants correlates positively with long term survival compared to the glass bead and electroporation methods.
Therefore, in order to express heterologous protein in N. salina, we first tested transformation efficiency when using particle bombardment. N. salina was transformed with plasmid expression constructs for the Shble marker gene under the control of the TUB or UEP promoter. The transformants were selected on agar plates containing Zeocin, and their transformation efficiencies were determined, as summarized in Table 2. These analyses showed that 5.9 and 4.7 transformants per 108 cells were obtained using TUB and UEP promoter constructs, respectively. These results are consistent with a previous report that used electroporation method for transformation of Nannochloropsis gaditana [5]. Here, it is worth to note that transformation efficiency can be improved by modification of transformation conditions, and that size and concentration of DNA fragment, which will be transferred into cells, affect the transformation efficiency. For instance, Li et al. suggested that transformation efficiency significantly increased, when using short DNA fragment for Nannochloropsis sp. transformation [7]. This is likely due to the fact that short DNA fragments improve their chances of integration. Therefore, an integrated development of bombardment based transformation is necessary to increase transformation efficiency.
Table 2.
Transformation efficiency of N. salina achieved by particle bombardment.
| Promoter | Colonies/108 cells | Colonies/μg DNA |
|---|---|---|
| TUB | 5.9 ± 1.6 | 5.9 ± 1.6 |
| UEP | 4.7 ± 2.0 | 4.7 ± 2.0 |
| No plasmida | 0 | 0 |
Data are means ± SD (n = 3).
No plasmid indicates a control bombardment transformation carried out without DNA.
To validate stable integration of the transgene in the genomic DNA and to identify the integration site, we employed RESDA PCR. The overall strategy and a vector map are shown in Fig. 1a. Fig. 1b shows the results of the second round of PCR (Amp II) using DegBamHI during RESDA PCR. The results obtained are consistent with a previous report that, in general, the average band size after Amp II ranges from 300 bp to 2 kb [14]. There were unique bands in many NsTShble transformant lines, denoted as ‘a’ through ‘d’. DNA from each band was then purified and sequenced to identify integration patterns because different band patterns represent different insertion sites of transgenes. Sequencing of one example transformant, NsTShble 2, revealed sequences of the transformed vector flanked by the genomic sequence of N. salina CCMP1776, in addition to the landmark primer sequences of S4, S5, and Q0 (Fig. 1c). Other identified sequences are shown in Fig. S1. Importantly, the size of each band and the length of the corresponding sequence coincided, indicating that the amplification and sequencing of the integration sites were correct. It has been recently reported that the organellar genomes of N. salina are more than 97% identical to genomes of N. gaditana [23]. Therefore, the flanking regions of each transformant were identified based on N. gaditana genome information [24], then we could indirectly estimate where transgenes were integrated as shown in Table S1.
The expression of heterologous protein is primarily influenced by the genomic environment in which the transgene is integrated. This is known as the position effect of transgenes [25]. Transgene silencing is the most devastating problem in genetic engineering; it can be caused by the position effect and/or by transcriptional and post-transcriptional silencing complexes identified in algae and plants [26], [27]. Such silencing effects can be counteracted using boundary elements called matrix attachment regions (MARs) to enhance transgene expression [28], as shown in Dunaliella salina [29]. Identification of the transgene integration sites in N. salina will help to clarify the integration patterns and could assist in the design of better strategies for achieving the stable expression of heterologous protein. For instance, we are able to find specific flanking sites resulting in better and stable expression of target protein through statistical analysis with lots of integration patterns of transformants.
Taken together, strategies combined with identification of stable integration sites and bombardment transformation is likely very useful for the production of stable transformants.
3.2. Heterologous overexpression of sfCherry fluorescent protein
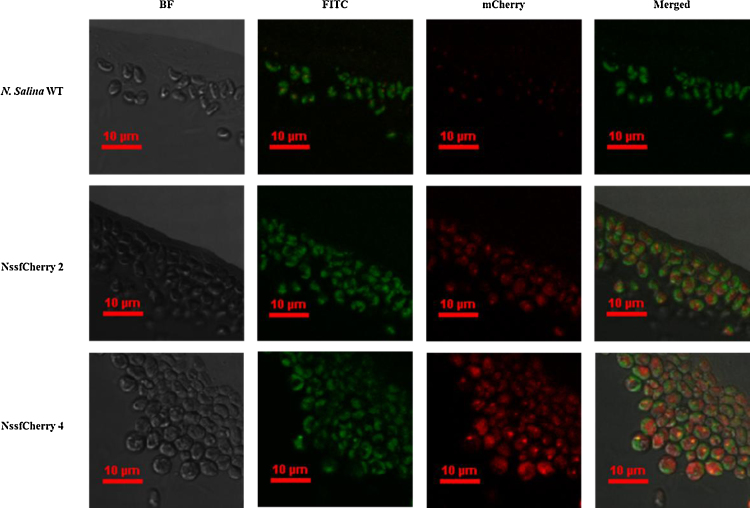
In order to assess the expression of a heterologous protein, we transformed N. salina with an expression construct for the gene encoding sfCherry fluorescent protein (Fig. 2a) and analyzed its expression by Western blot analysis and confocal microscopy. Transformant candidates were initially analyzed by PCR using S1 and S2 primers (Table 1) to amplify part of the Shble gene as well as the positive control, 18S rDNA (Fig. 2b and c). PCR-positive candidates were subsequently analyzed for expression of the sfCherry fluorescent protein by western blotting. These analyses showed the expression of a protein with a molecular weight of about 27 kD, consistent with the expected size of 27.39 kD based on the mCherry amino acid sequence (Fig. 2d). To further validate the expression of functional sfCherry fluorescent protein, we analyzed transformants NssfCherry 2 and 4, which Western blot analyses showed were strongly positive for sfCherry, using confocal fluorescence microscopy (Fig. 3). FITC fluorescence (auto-fluorescence) was detected in both wild-type controls and transformants, but only transformants expressed sfCherry-specific fluorescence (excitation: 561 nm, emission: 595/50 nm). The intensity of sfCherry fluorescence in NssfCherry 4 was stronger than that of NssfCherry 2. This is likely due to the position effect, which can affect the expression level of fluorescent protein [25], [28]. These results indicate that particle bombardment can stably deliver DNA into N. salina cells, and produce correctly folded functional proteins.
Fig. 3.
Confocal microscopic images of NssfCherry 2 and 4 compared with wild-type control. First column: bright field (BF) images; second column: FITC filter images showing autofluorescence; third column: images obtained with the sfCherry filter. Merged images of the three channels are shown in the last column.
Fluorescence proteins have been used for structural and functional studies in various cell lines [30]. In addition, there is an increasing trend toward the application of flow cytometry and fluorescent markers, which have been available for use with other organisms as well as microalgae [31]. We previously transformed N. salina with expression constructs for cyan fluorescence protein (CFP), but were unable to detect CFP fluorescence signal owing to the high background (Fig. S2), most likely reflecting the cellular pigments present in Nannochloropsis. In contrast, N. salina cells were successfully transformed and confirmed to express functional sfCherry fluorescent protein without interference from endogenous pigments. sfCherry was developed by directed mutagenesis for efficient folding and strong fluorescence [17], [32]; thus, this protein has great potential for the structural and functional analyses necessary for the genetic engineering of Nannochloropsis. In particular, sfCherry fluorescent protein can be applied for development of selection marker, monitoring the real time expression level of fusion protein, and imaging of the localization and dynamics of specific organelles [15].
Furthermore, because microalgal chloroplasts can correctly fold complex proteins, microalgae, especially Chlamydomonas reinhardtii, represent an emerging source for the production of various bioproducts, such as nutraceuticals and therapeutics [11], [33]. Another attractive feature of C. reinhardtii as a heterologous protein factory is that a relatively short time is required to yield bioproducts [12]. Although Nannochloropsis will still require genetic improvements to be used as a strain for the production of biofuels and bioproducts such as EPA, our demonstration of heterologous protein expression in Nannochloropsis suggests the possibility of this strain as a cell factory for biofuels and bioproducts.
4. Conclusions
Heterologous protein, sfCherry fluorescent protein, was overexpressed and visualized in N. salina for the first time. To do transformation, particle bombardment was employed, and TUB and UEP promoters were used to express the Shble gene, yielding 5.9 and 4.7 transformants per 108 cells, respectively. Genomic integration, confirmed by RESDA PCR, ensured the stable expression of the transgenes. Furthermore, transgenic expression and correct function of the sfCherry fluorescent protein were confirmed by western blotting and confocal microscopy. These results provide techniques for the genetic manipulation of Nannochloropsis that may be useful for the stable transformation and production of bioproducts.
Acknowledgments
We thank Dr. Won Joong Jeong at KRIBB for technical support, including Gene Gun applications. This work was supported by the Advanced Biomass R&D Center (ABC) of Global Frontier Project funded by the Ministry of Science, ICT and Future Planning (ABC-2010-0029728 and 2011-0031350).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2015.08.004.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Yen H.W., Hu I.C., Chen C.Y., Ho S.H., Lee D.J., Chang J.S. Microalgae-based biorefinery – from biofuels to natural products. Bioresour. Technol. 2013;135:166–174. doi: 10.1016/j.biortech.2012.10.099. [DOI] [PubMed] [Google Scholar]
- 2.Williams P.J.l.B., Laurens L.M.L. Microalgae as biodiesel & biomass feedstocks: review & analysis of the biochemistry, energetics & economics. Energy Environ. Sci. 2010;3:554. [Google Scholar]
- 3.Wang D.M., K. Ning J., Li J.Q., Hu D.X., Han H., Wang X.W., Zeng X.Y., Jing Q., Zhou X.Q., Su X.Z., Chang A.H., Wang W., Wang J., Jia L., Wei Y., Xin Y.H., Qiao R.R., Huang J., Chen B., Han K., Yoon R.T., Hill Y., Zohar F., Chen Q. Hu. Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 2014;10:e1004094. doi: 10.1371/journal.pgen.1004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal D., Khozin-Goldberg I., Cohen Z., Boussiba S. The effect of light, salinity, and nitrogen availability on lipid production by Nannochloropsis sp. Appl. Microbiol. Biotechnol. 2011;90:1429–1441. doi: 10.1007/s00253-011-3170-1. [DOI] [PubMed] [Google Scholar]
- 5.Radakovits R., Jinkerson R.E., Fuerstenberg S.I., Tae H., Settlage R.E., Boore J.L., Posewitz M.C. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 2012;3:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieler A., G. Wu C.H., Tsai B., Bullard A.J., Cornish C., Harvey I.B., Reca C., Thornburg R., Achawanantakun C.J., Buehl M.S., Campbell D., Cavalier K.L., Childs T.J., Clark R., Deshpande E., Erickson A. Armenia Ferguson, Handee W., Kong Q., Li X., Liu B., Lundback S., Peng C., Roston R.L., Sanjaya J.P., Simpson A., Terbush J., Warakanont S., Zauner E.M., Farre E.L., Hegg N., Jiang M.H., Kuo Y., Lu K.K., Niyogi J., Ohlrogge K.W., Osteryoung Y. Shachar-Hill, Sears B.B., Sun Y., Takahashi H., Yandell M., S.H. Shiu C. Benning. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 2012;8:e1003064. doi: 10.1371/journal.pgen.1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., D. Gao H. Hu. High-efficiency nuclear transformation of the oleaginous marine Nannochloropsis species using PCR product. Biosci. Biotechnol. Biochem. 2014;78:812–817. doi: 10.1080/09168451.2014.905184. [DOI] [PubMed] [Google Scholar]
- 8.Kang N.K., Lee B., Shin S.E., Jeon S., Park M.S., Yang J.W. Use of conditioned medium for efficient transformation and cost-effective cultivation of Nannochloropsis salina. Bioresour. Technol. 2015;181:231–237. doi: 10.1016/j.biortech.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 9.Kilian O., Benemann C.S., Niyogi K.K., Vick B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21265–21269. doi: 10.1073/pnas.1105861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coll J.M. Methodologies for transferring DNA into eukaryotic microalgae. Span. J. Agric. Res. 2006;4:316–330. [Google Scholar]
- 11.Tran M., Van C., Barrera D.J., Pettersson P.L., Peinado C.D., Bui J., Mayfield S.P. Production of unique immunotoxin cancer therapeutics in algal chloroplasts. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E15–E22. doi: 10.1073/pnas.1214638110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayfield S.P., A.L. Manuell S., Chen J., Wu M., Tran D., Siefker M. Muto. Chlamydomonas reinhardtii chloroplasts as protein factories. Curr. Opin. Biotechnol. 2007;18:126–133. doi: 10.1016/j.copbio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y.G. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Ballester D., de Montaigu A., Galvan A., Fernandez E. Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal. Biochem. 2005;340:330–335. doi: 10.1016/j.ab.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 15.Giepmans B.N., Adams S.R., Ellisman M.H., Tsien R.Y. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 16.Shaner N.C., Steinbach Tsien P.A.R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen H.B., Hung L.W., Yeates T.O., Terwilliger T.C., Waldo G.S. Split green fluorescent protein as a modular binding partner for protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 2013;69:2513–2523. doi: 10.1107/S0907444913024608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillard R.R., Ryther J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 19.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison 3rd C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 20.Lim J.M., Ahn J.W., Hwangbo K., Choi D.W., Park E.J., Hwang M.S., Liu J.R., Jeong W.J. Development of cyan fluorescent protein (CFP) reporter system in green alga Chlamydomonas reinhardtii and macroalgae Pyropia sp. Plant Biotechnol. Rep. 2013;7:407–414. [Google Scholar]
- 21.Nakayama T., Watanabe S., Mitsui K., Uchida H., Inouye I. The phylogenetic relationship between the Chlamydomonadales and Chlorococcales inferred from 18SrDNA sequence data. Phycol. Res. 1996;44:47–55. [Google Scholar]
- 22.Cerutti H., Johnson A.M., Gillham N.W., Boynton J.E. Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell. 1997;9:925–945. doi: 10.1105/tpc.9.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starkenburg S.R., Kwon K.J., Jha R.K., McKay C., Jacobs M., Chertkov O., Twary S., Rocap G., Cattolico R.A. A pangenomic analysis of the Nannochloropsis organellar genomes reveals novel genetic variations in key metabolic genes. BMC Genomics. 2014;15:212. doi: 10.1186/1471-2164-15-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corteggiani Carpinelli E., Telatin A., Vitulo N., Forcato C., D’Angelo M., Schiavon R., Vezzi A., Giacometti G.M., Morosinotto T., Valle G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant. 2013 doi: 10.1093/mp/sst120. [DOI] [PubMed] [Google Scholar]
- 25.Matzke A.J., Matzke M.A. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1998;1:142–148. doi: 10.1016/s1369-5266(98)80016-2. [DOI] [PubMed] [Google Scholar]
- 26.Wu-Scharf D., Jeong B., Zhang C., Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- 27.Jeong Br B.R., Wu-Scharf D., Zhang C., Cerutti H. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1076–1081. doi: 10.1073/pnas.022392999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen G.C., Spiker S., Thompson W.F. Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol. Biol. 2000;43:361–376. doi: 10.1023/a:1006424621037. [DOI] [PubMed] [Google Scholar]
- 29.Wang T., L. Xue W., Hou B., Yang Y., Chai X. Ji. Increased expression of transgene in stably transformed cells of Dunaliella salina by matrix attachment regions. Appl. Microbiol. Biotechnol. 2007;76:651–657. doi: 10.1007/s00253-007-1040-7. [DOI] [PubMed] [Google Scholar]
- 30.Chudakov D.M., Matz M.V., Lukyanov S., Lukyanov K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 31.Hyka P., Lickova S., Pribyl P., Melzoch K., Kovar K. Flow cytometry for the development of biotechnological processes with microalgae. Biotechnol. Adv. 2013;31:2–16. doi: 10.1016/j.biotechadv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 33.Bertalan I., Munder M.C., Weiss C., Kopf J., Fischer D., Johanningmeier U. A rapid, modular and marker-free chloroplast expression system for the green alga Chlamydomonas reinhardtii. J. Biotechnol. 2015;195:60–66. doi: 10.1016/j.jbiotec.2014.12.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.