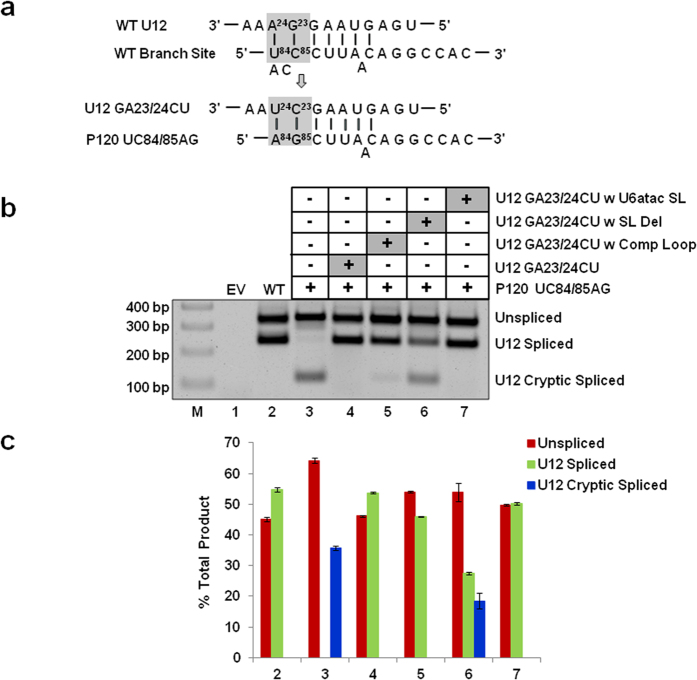
Figure 2. Effect of replacing the SLIII in U12 snRNA with the U6atac distal 3′ SL on in vivo splicing.
(a) Features of the branch site suppression assay. Wild type (WT) base pairing between human U12 snRNA and the branch site of the U12-dependent intron of P120 pre-mRNA is shown. The boxed nucleotides were mutated to their complementary nucleotides as shown. GA nucleotides at positions 23/24 in U12 snRNA were mutated to CU and the corresponding nucleotides UC at positions 84/85 in the branch site were mutated to AG. U12 GA23/24CU mutations are required to fully suppress the effect of the branch site UC84/85AG mutation. (b) Splicing phenotypes of P120 WT and the P120 UC84/85AG mutant coexpressed with the indicated U12 snRNA mutants. CHO cells were transfected with the indicated constructs and splicing phenotypes were assayed by RT–PCR. Lane M: 100 bp ladder. U12 GA23/24CU w U6atac SL denotes the U12 GA23/24CU snRNA construct containing the U6atac distal 3′ SL in place of U12 SLIII. The positions of bands corresponding to unspliced RNA, RNA spliced at the normal U12-dependent splice sites (U12 spliced) and RNA spliced at the cryptic U12-dependent splice sites (U12 cryptic spliced) are indicated. The cryptic spliced product results from the activation of a U12-dependent cryptic splice site in the downstream exon. (c) Quantitative analysis of spliced/unspliced products. Numbers (x-axis) correspond to the respective lanes of the gel shown in (b). Error bars represent ± SE of three experiments.