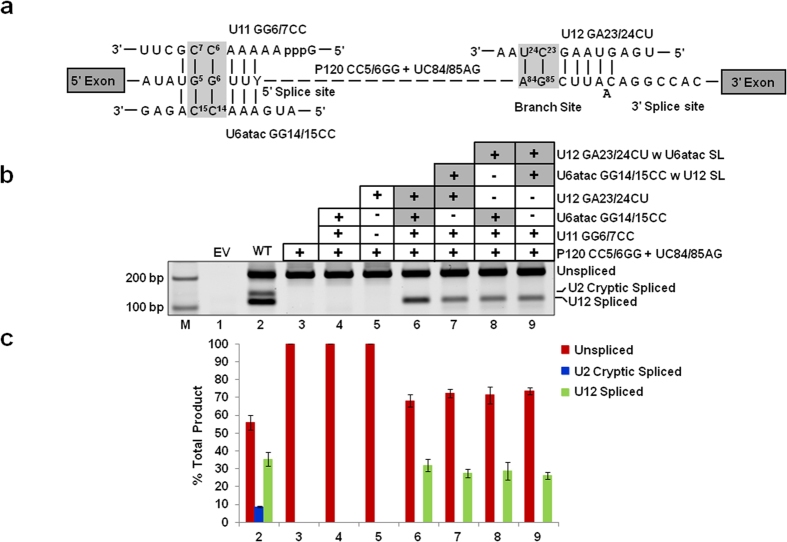
Figure 4. Combined effect of the exchange of SLs in both U6atac and U12 snRNAs on in vivo splicing.
(a) Features of the binary splice site suppression assay. In this assay, both branch site and 5′ SS mutations shown in Figs 2 and 3 were present in the P120 U12-dependent intron. The U12-dependent intron containing 5′ SS CC5/6GG and branch site UC84/85AG mutations is shown. The base pairing interactions between U11 GG6/7CC, U6atac GG14/15CC, U12 GA23/24CU snRNAs and the mutant 5′ SS and branch site of P120 intron are also shown. Boxed nucleotides denote the mutated nucleotides as shown in Figs 2 and 3. (b) Splicing phenotypes of P120 WT and the P120 CC5/6GG + UC84/85AG mutant coexpressed with the indicated suppressor snRNA constructs. CHO cells were transiently transfected with the indicated constructs and total RNA was extracted. The splicing pattern of the U12-dependent P120 intron was analyzed by RT-PCR using primers designed to bind flanking exons. Lane M: 100 bp ladder. The positions of bands corresponding to unspliced RNA, RNA spliced at the normal U12-dependent splice sites (U12 spliced) and RNA spliced at the cryptic U2-dependent splice sites (U2 cryptic) are indicated. (c) Quantitative analysis of spliced/unspliced products. Numbers on the x-axis correspond to the respective lanes of the gel shown in (b). Error bars represent ± SE of three experiments.