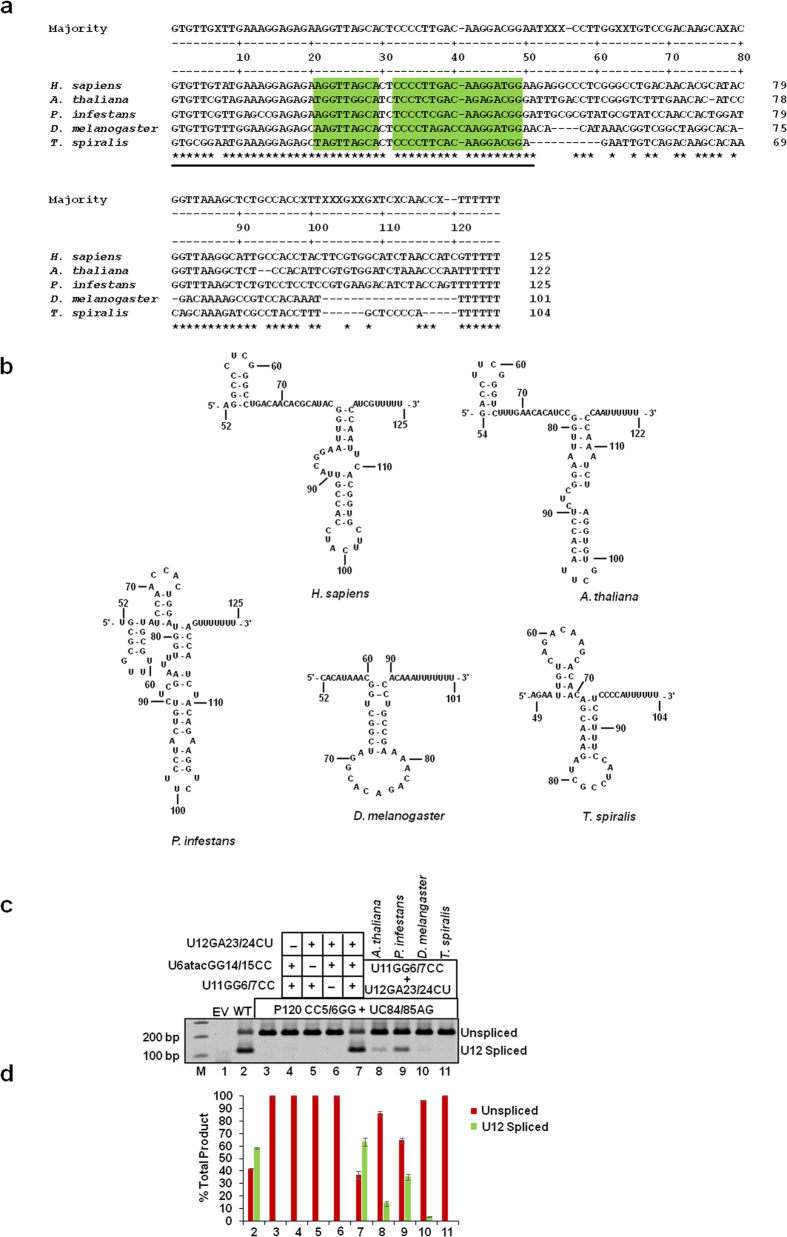
Figure 5. Activity of chimeric U6atac snRNAs in U12-dependent splicing in vivo.
(a) Comparative sequence analysis of U6atac snRNAs from the indicated species. ‘*’ denotes the conservation of a nucleotide in at least three species. Nucleotide sequences in green boxes represent stem I and stem II formed by RNA-RNA base pair interactions between U4atac:U6atac snRNAs. The region of U6atac snRNA denoted by a solid line below the consensus nucleotide sequences at the 5′ end of U6atac snRNA is referred to as the “business end” of the molecule. (b) Predicted MFold secondary structures of the 3′ end of U6atac snRNA from Homo sapiens, Arabidopsis thaliana, Phytophthora infestans, Drosophila melanogaster and Trichnella spiralis. (c) Splicing phenotypes of the binary splice site mutant (P120 CC5/6GG + UC84/85AG) coexpresssed with human chimeric U6atac snRNA containing the 3′ end of U6atac snRNA from the species shown in Panel a & b. Transfections, RT-PCR, cDNA amplification and gel electrophoresis was performed as described in the legend to Fig. 4. (d) Quantitation of unspliced and spliced products using Image J software. Numbers (x-axis) correspond to the respective lanes of the gel shown in (c).