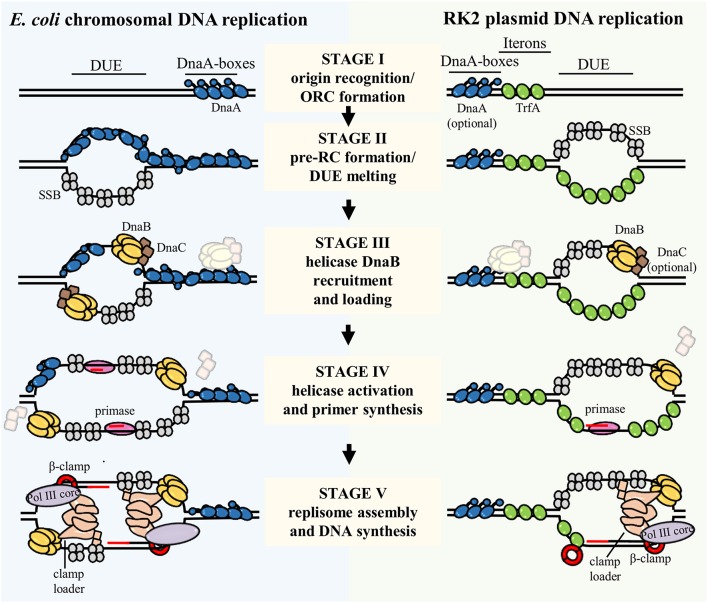
Figure 2.
The process of bacterial chromosome and plasmid DNA replication initiation and replisome assembly. The scheme presents replication initiation and replisome assembly at chromosomal E. coli origin, oriC (left), and RK2 plasmid origin, oriV (right). The DNA replication initiation starts with binding a replication initiator(s) DnaA and TrfA to the DnaA boxes and Iterons, respectively (Stage I). Origin Recognition Complex (ORC) formation induces local destabilization and pre-Replication Complex (pre-RC) formation and melting of the DNA Unwinding Element (DUE) region (Stage II). Then, assisted by replication initiators and the DnaC helicase loader, the DnaB helicase is recruited and loaded onto the single-stranded DUE (Stage III). In case of plasmid DNA replication the requirement for DnaA and DnaC is optional as it depends on the host organism. Association of DnaG primase triggers the release of helicase loader, helicase activation and primers synthesis (Stage IV). Next, the holoenzyme of DNA Polymerase III, which comprises clamp loader, DNA Polymerase III core (Pol III core), and β-clamp is assembled and conducts DNA synthesis (Stage V). Lagging strand synthesis was omitted for simplicity. Proteins involved in described stages of DNA replication initiation and replisome assembly processes are depicted in the scheme. IHF and Fis were omitted in this scheme.