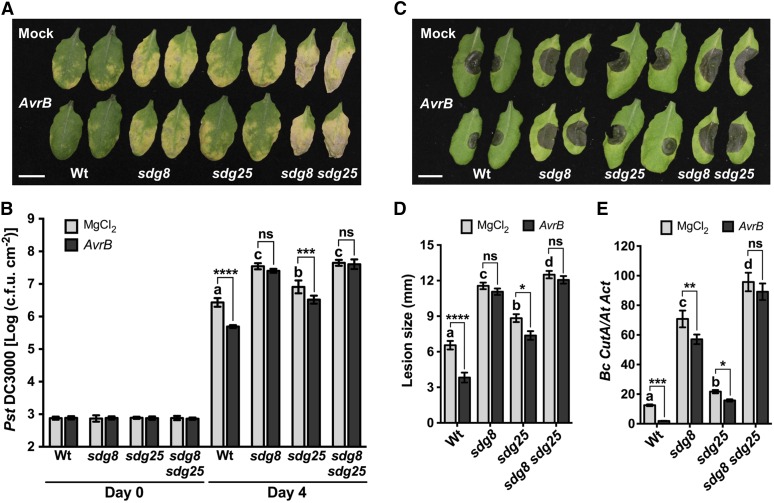
Figure 2.
SDG8 and SDG25 Are Required for SAR to Bacterial and Fungal Pathogens.
(A) and (B) Impaired systemic resistance of sdg mutants to bacterial infection. Disease symptoms (A) at 4 d after bacterial inoculation and bacterial growth (B) in secondary leaves at 0 and 4 d after inoculation with the virulent bacterial strain Pst DC3000 with 10 mM MgCl2. Data represent mean log (cfu/cm2), and error bars indicate ± sd from six replicates. Bar = 1 cm.
(C) to (E) Impaired systemic resistance of sdg mutants to fungal infection. Disease symptoms (C), disease lesion size (D), and fungal growth (E) in secondary leaves at 3 d after drop inoculation with B. cinerea. Fungal growth was determined by qPCR amplification of the B. cinerea Cutinase DNA (Bc CutA). The relative DNA levels were calculated by the comparative cycle threshold method with Arabidopsis Actin2 as the reference gene. Bar = 1 cm.
Data in (D) represent means ± se (n ≥ 39 disease lesions) and in (E) represent mean ± sd (n = 9). In all cases, Pst DC3000 (OD600 = 0.001) or B. cinerea (2.5 × 105 spores/mL) were inoculated on secondary leaves 2 d after lower leaves were inoculated with 10 mM MgCl2 or avirulent strain Pst DC3000 carrying AvrB (OD600 = 0.02) to activate SAR. Asterisks indicate significant difference between mock and AvrB treatments (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; two-way ANOVA). Different letters indicate significant differences among genotypes (P < 0.05, Student’s t test). The experiments were repeated at least three times with similar results. Mock, plants pretreated with 10 mM MgCl2; Wt, wild type; sdg8, sdg8-2; sdg25, sdg25-1; ns, no significance.