Figure 1.
Different Catalytic Roles of the Four Mg2+-Coordinating Active-Site Residues.
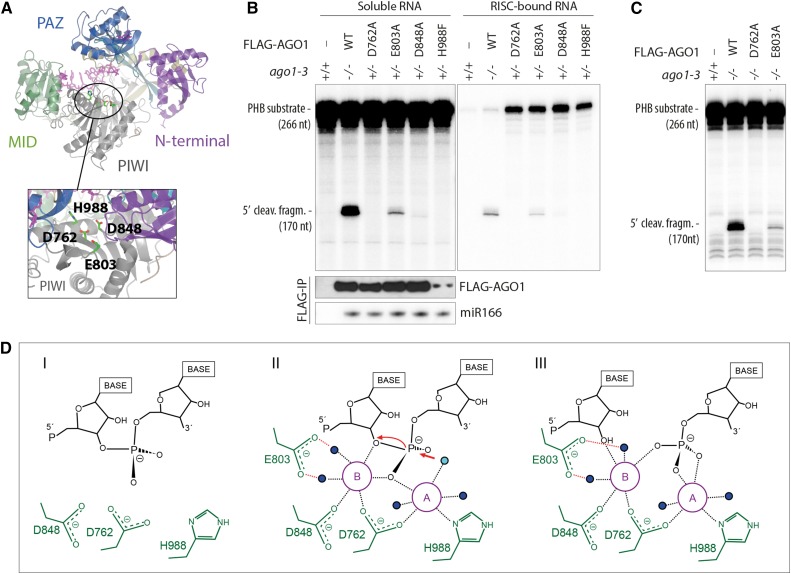
(A) Structural model of AGO1 based on the crystal structure of human Ago2 (PDB ID 4EI3). The locations of the four residues involved in coordination of Mg2+-ions are indicated.
(B) Slicer activity of immuno-affinity-purified FLAG-AGO1 from Arabidopsis inflorescences of stable transgenic lines expressing wild-type and slicer-deficient FLAG-AGO1. A 266-nucleotide 32P cap-labeled PHB single-stranded RNA bearing the endogenous miR166 target site was used as substrate. FLAG-AGO1 was immobilized on M2-conjugated beads during the reaction. The left panel shows RNA extracted from the supernatant of the reaction. The right panel shows RNA extracted from the agarose beads that contain immobilized FLAG-AGO1. Cleavage activity is revealed by the accumulation of the 5′ cleavage fragment. Nontransgenic Col-0 was used to control for unspecific binding of endogenous AGO1 to M2-conjugated beads (left lanes). A reproduction of this panel with higher contrast can be found in Supplemental Figure 1 to facilitate visualization of the 5′-cleavage fragment produced by FLAG-AGO1D848A. FLAG IPs were split into three parts for analysis of cleavage activity, FLAG-AGO1 levels and miR166 levels. Lower panels show the amount of FLAG-AGO1 and miR166 purified in the IP and used in the slicer assay by protein and RNA gel blot analyses, respectively. The weak FLAG signal for FLAG-AGO1H988F detected on this membrane was in part due to sample loss during gel loading and was not observed in other experiments (see Supplemental Figure 1 for an example). This is consistent with the nearly equal amount of miR166 immunoprecipitated from FLAG-AGO1H988F compared with the other lines. Additional controls show that slicer activity is detectable in less than 1% of the amount of FLAG-AGO1WT used here (Supplemental Figure 1), corroborating the conclusion that FLAG-AGO1H988F is defective in slicer activity.
(C) Slicer activity of immuno-affinity-purified FLAG-AGO1 from Arabidopsis seedlings of stable transgenic lines expressing wild-type and slicer-deficient FLAG-AGO1 in an ago1-3 homozygous background. The assay was performed as in (B), and RNA was extracted from the whole reaction.
(D) Proposed slicing mechanism of Arabidopsis AGO1. Steps in RNA cleavage (I, II, and III) follow the previously proposed structure-based model for slicing by Thermus thermophilus Ago (Sheng et al., 2014). (I) Precleavage state: Three active site residues in close proximity to the scissile phosphate bond of the RNA target. Absence of the glutamic finger (E803) and probably also of metal ions. (II) Cleavage state: An activated water molecule (cyan) coordinated by Mg2+ ion A (magenta) is positioned to perform a nucleophile attack on the scissile phosphodiester. Both ions A and B are involved in coordinating the pentavalent hydrolysis intermediate. Note the presence of the glutamic finger and its role in coordinating metal ion B through two water molecules. (III) Postcleavage state before product release: All active-site residues and metal ions are present. Water molecules not directly involved in catalysis are colored blue.