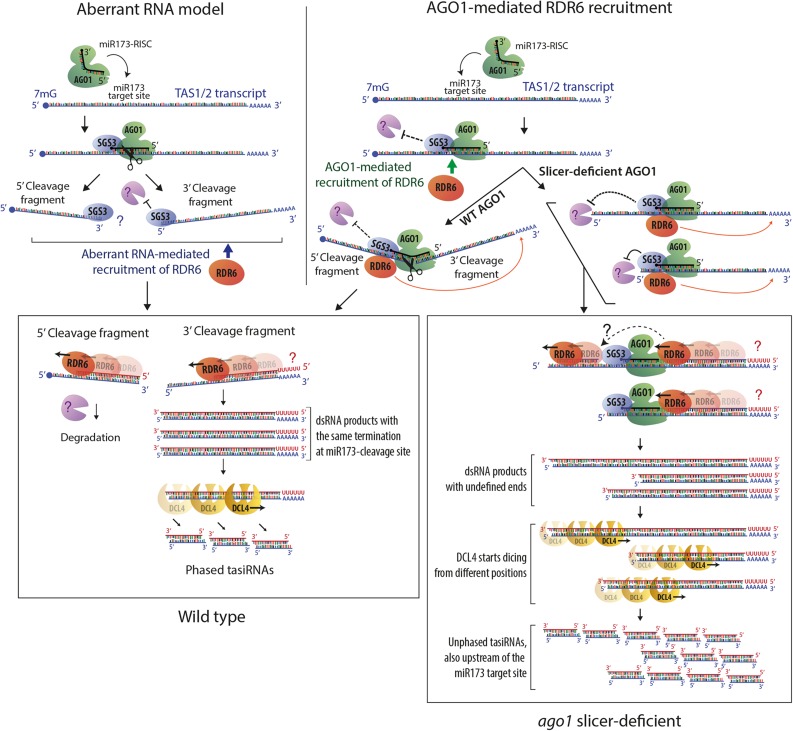
Figure 7.
Models of RDR6 Recruitment to TAS Transcripts.
Schematic diagram highlighting main features of two models to explain RDR6 recruitment to TAS transcripts and the function of SGS3. In the aberrant RNA model (left), cleavage of TAS precursors is required to produce RNA fragments with aberrant features. SGS3 binds to the miR173/TAS duplex region protruding from RISC, and the role of SGS3 is to stabilize cleavage fragments to provide substrate for RDR6 (Fukunaga and Doudna, 2009; Yoshikawa et al., 2013). Protection of the 5′-cleavage fragment by SGS3, and RDR6-mediated conversion into dsRNA for degradation is postulated because steady state levels of the 5′-cleavage fragment increase substantially in rdr6 mutants and disappear in sgs3 mutants (Yoshikawa et al., 2005). In the model on the right, the AGO1 protein is required to recruit RDR6, and its miR173-guided precursor cleavage ensures phasing because it generates well-defined 5′-ends of the RDR6 substrate. This model readily explains how tasiRNAs are generated in catalytic AGO1 mutants. The model assumes that two mechanisms may contribute to preferential accumulation of siRNAs 3′ to the miR173 site: A road block function of bound AGO1/SGS3 limits RDR6 processivity to the miR173 site in most cases (Rajeswaran and Pooggin, 2012) and stops 5′-3′-exonucleolysis of those transcripts that escape protection against degradation provided by bound SGS3. On transcripts in which AGO1/SGS3 dissociate, RDR6 continues until the 5′-end, consistent with dsRNA mapping performed by Rajeswaran et al. (2012). Together, these processes give rise to a pool of DCL substrate dsRNAs with different ends, thus explaining the unphased nature of tasiRNAs in catalytic ago1 mutants. In both models, RDR6 is shown to initiate at the end of poly(A) tails, but the question marks indicate that it is unclear whether deadenylated or polyadenylated RNAs are used as templates in vivo.