Cell cycle-regulated CSLD5 provides essential cell wall synthase activity for cell plate formation during cytokinesis.
Abstract
In plants, the presence of a load-bearing cell wall presents unique challenges during cell division. Unlike other eukaryotes, which undergo contractile cytokinesis upon completion of mitosis, plants instead synthesize and assemble a new dividing cell wall to separate newly formed daughter cells. Here, we mine transcriptome data from individual cell types in the Arabidopsis thaliana stomatal lineage and identify CSLD5, a member of the Cellulose Synthase Like-D family, as a cell wall biosynthesis enzyme uniquely enriched in rapidly dividing cell populations. We further show that CSLD5 is a direct target of SPEECHLESS, the master transcriptional regulator of these divisions during stomatal development. Using a combination of genetic analysis and in vivo localization of fluorescently tagged fusion proteins, we show that CSLD5 preferentially accumulates in dividing plant cells where it participates in the construction of newly forming cell plates. We show that CSLD5 is an unstable protein that is rapidly degraded upon completion of cell division and that the protein turnover characteristics of CSLD5 are altered in ccs52a2 mutants, indicating that CSLD5 turnover may be regulated by a cell cycle-associated E3-ubiquitin ligase, the anaphase-promoting complex.
INTRODUCTION
In multicellular organisms, development and differentiation is associated with successive rounds of cell division in self-renewing populations of embryonic and postembryonic stem cells (Heidstra and Sabatini, 2014). Unlike most other eukaryotes, which undergo contractile cytokinesis to separate daughter cells upon completion of mitosis (Guertin et al., 2002), plants instead deposit a new dividing cell wall, which is formed across the plane of division and separates the two daughter cells (Jürgens, 2005; Inagaki and Umeda, 2011). The construction of this new cell wall, requiring rapid synthesis and delivery of plant cell wall polysaccharides to the newly forming cell plate, represents a novel and unique process associated with cytokinesis in plants (Hong et al., 2001; Yokoyama and Nishitani, 2001; Miart et al., 2014).
The major load-bearing component in plant cell walls is cellulose, which is made by plasma membrane-localized cellulose synthases, called CESA proteins (Cosgrove, 2005). In plants, CESA proteins share significant sequence similarity to a larger set of proposed glycan synthases, called the Cellulose Synthase-Like (CSL) gene family (Richmond and Somerville, 2001). Within the Arabidopsis thaliana CSL superfamily, the Cellulose Synthase Like-D family (CSLD) displays a high degree of sequence identity with CESA sequences, containing extended amino terminal and expanded catalytic domains, which discriminate these groups from other CSL families. Isolation of root hairless csld3 mutants (Favery et al., 2001; Wang et al., 2001) implicated this class of cell wall synthases in tip-restricted cell expansion. Subsequent demonstration that csld2 mutants also displayed root hair defects and specific roles for CSLD1 and CSLD4 in pollen, another tip-growing cell type, further supported important roles for CSLDs during cell wall synthesis in tip-growing cells (Bernal et al., 2008). However, mutation of CSLD5 does not result in defective tip growth (Bernal et al., 2007). This, combined with the observation that csld2 csld5 (csld2/5) or csld3 csld5 (csld3/5) double mutants display dramatic growth defects, supports broader roles for CSLD proteins than cell wall deposition in tip-growing cells. Initial studies had proposed roles for CSLD proteins in the synthesis of xylan, homogalacturonan, or mannan polysaccharides (Bernal et al., 2007; Yin et al., 2011; Verhertbruggen et al., 2011). More recently, functional fluorescent CSLD3 fusion proteins were detected in apical plasma membranes of growing root hairs, a region where high levels of new cellulose synthesis was observed (Park et al., 2011; Galway et al., 2011), and rescue of csld3 root hair phenotypes by a chimeric CSLD3 fusion protein containing the catalytic domain of CESA6 supports the possibility that at least some members of the CSLD family, such as CSLD3, might also provide β-1,4-linked glucan synthase activity (Park et al., 2011).
Because cellulose synthesis is required for growth and division of all cells, mutations in CESA and CSL genes are often pleiotropic, and because the gene families are large, redundancy has often masked the roles of individual genes. One approach to bypass these issues is to take advantage of specific cell types or growth conditions that promote cell expansion or division. Such approaches identified roles for CESA6 (PROCUSTE) during hypocotyl cell elongation (Fagard et al., 2000) and linked the function of CSLD3 with the synthesis and deposition of cellulose-like cell wall polysaccharides in apical plasma membranes during root hair tip growth (Park et al., 2011).
Here, we mine transcriptome data from individual cell types in the stomatal lineage, which represent proliferative, self-renewing, and differentiating cell types, to identify CSLD5 as a cell wall biosynthesis enzyme uniquely enriched in the self-renewing meristemoid population (Adrian et al., 2015). We further show that CSLD5 is a direct target of SPEECHLESS (SPCH), the master transcriptional regulator of these divisions (Lau et al., 2014). Using a combination of genetic analysis and in vivo localization of fluorescently tagged fusion proteins, we show that CSLD5 preferentially accumulates in dividing plant cells, where it localizes to and participates in the synthesis of newly forming cell plates. In addition, we show that CSLD5, unlike other CSLD family members, and the closely related CESA family of cellulose synthases, is an unstable protein that is rapidly degraded upon completion of cell division. Finally, we show that the protein turnover characteristics of CSLD5 are altered in ccs52a2 mutants, indicating that CSLD5 protein turnover may be regulated by the cell cycle-associated E3-ubiquitin ligase, the anaphase-promoting complex (APC).
RESULTS
Disruption of CSLD5, a SPCH Target, Impairs Stomatal Patterning
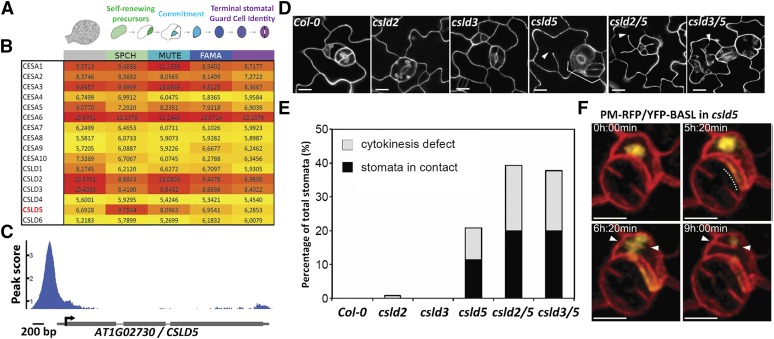
Plant cytokinesis involves the de novo construction of new cell walls in membrane-bound phragmoplast compartments that eventually fuse with the plasma membrane. We reasoned that plasma membrane-targeted CESA or CSLD families would be required for this process but that different members might be required for division and expansion. To enrich for genes expressed in dividing cells, we took advantage of recent transcriptional profiles of the stomatal lineage, which separate self-renewing cells that primarily undergo asymmetric cell divisions, differentiating guard cells, and expanding pavement cell populations (Adrian et al., 2015) (Figure 1A). We found that Arabidopsis CESA genes associated with the formation of cellulose in primary, but not secondary, cell walls were highly expressed in both developing leaf epidermal cells and in stomatal stem cell lineages, which would be consistent with roles in cell expansion observed in these tissues (Figure 1B). Additionally, three CSLD genes expressed in vegetative Arabidopsis tissues (Bernal et al., 2008), CSLD2, CSLD3, and CSLD5, were also highly expressed (Figure 1B). However, unlike CESA genes, CSLD2, CSLD3, and CSLD5 displayed a distinct expression profile; high levels of expression were primarily observed in early-stage stomatal lineage cells and CSLD5 was largely absent from expanding and differentiating cells (Figure 1B). Consistent with this, CSLD5 was identified to be a direct target of the early stomatal lineage transcription factor SPCH. Chromatin immunoprecipitation (ChIP) with SPCHpro:SPCH-Myc revealed binding of this transcription factor to proximal regions in the CSLD5 5′ regulatory region (Figure 1C), and CSLD5 expression was upregulated upon SPCH induction (Lau et al., 2014).
Figure 1.
CSLD5 Is Specifically Enriched in Dividing Stomatal Cells and Required for Stomatal Cell Division.
(A) Diagram of stomatal lineage cell types from which FACS-isolated populations were transcriptionally profiled (adapted from Adrian et al., 2015).
(B) Heat map of regularized and transformed (via DESeq2) log2 expression values (RNA-seq) of cell wall biosynthesis genes CesA1-10 and CSLD1-6 during stomatal lineage progression (red = highest expression, yellow = lowest; expression level ranges from 0 to 20; genes with expression values below 3 are typically not detected by reporter or microarray); CSLD5 is uniquely enriched in asymmetrically dividing meristemoids.
(C) ChIP-seq trace of SPCHpro:SPCH-Myc binding to the 5′ regulatory region of CSLD5.
(D) Confocal images of leaf epidermis 5 d after initiation cotyledons, stained with propidium iodide, showing stomatal patterning and division defects (arrowheads) in csld single and double mutants.
(E) Quantification of stomatal defects shown in (D) (n ≥ 4 leaves from ≥ 4 individuals per genotype).
(F) Time-lapse images of cells expressing a PM-RFP marker protein (mCherry-RCl2A) and the polarity protein YFP-BASL, which is correctly localized despite division defects in csld5. Dashed line indicates polarized YFP-BASL accumulation, and arrowheads indicate incomplete cell plates. Bars = 10 µm in (D) and (F).
To test whether the enriched CSLD5 expression was associated with specific functions in stomatal development, we examined the stomatal lineages of 5- to 7-d-old csld2, csld3, and csld5 mutants. While leaf epidermal pavement cells appeared normal, csld5 mutant plants displayed significantly increased numbers of clustered stomatal guard cells (Figures 1D and 1E), and incomplete cell walls were observed in developing stomatal guard cell complexes (Figure 1D). Single csld2 and csld3 mutants did not display stomatal defects, but loss of either gene enhanced the csld5 phenotype (Figures 1D and 1E), consistent with a previous report showing the functional redundancy among CSLD genes (Yin et al., 2011). Since loss of CSLD5 activity interfered with normal stomatal development, often resulting in clustered stomatal guard cell complexes that resembled phenotypes observed in BASL (BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE) mutants (Dong et al., 2009), we monitored cell polarity using a functional BASL-fluorescent protein fusion during stomatal development in csld5 mutant seedlings (Figure 1F; Supplemental Movie 1). Intriguingly, cell polarization did not appear to be significantly impaired in csld5 mutants, and normal cortical polarization of YFP-BASL could be observed (Figure 1F, white bar in second panel), even during cell division events that resulted in the formation of incomplete cell walls (Figure 1F, arrowheads). Therefore, increased numbers of clustered stomatal guard cell complexes observed in csld5, csld2 csld5 (csld2/5), and csld3 csld5 (csld3/5) mutants may be primarily due to the cytokinesis defects associated with loss of csld function. These results suggest that the stomatal lineage provides a sensitized background in which to uncover CSLD5 activity at a cellular level but that CSLD5 activity may be required in all dividing cells.
CSLD5 Is Required for Proper Cell Wall Deposition during Cell Division
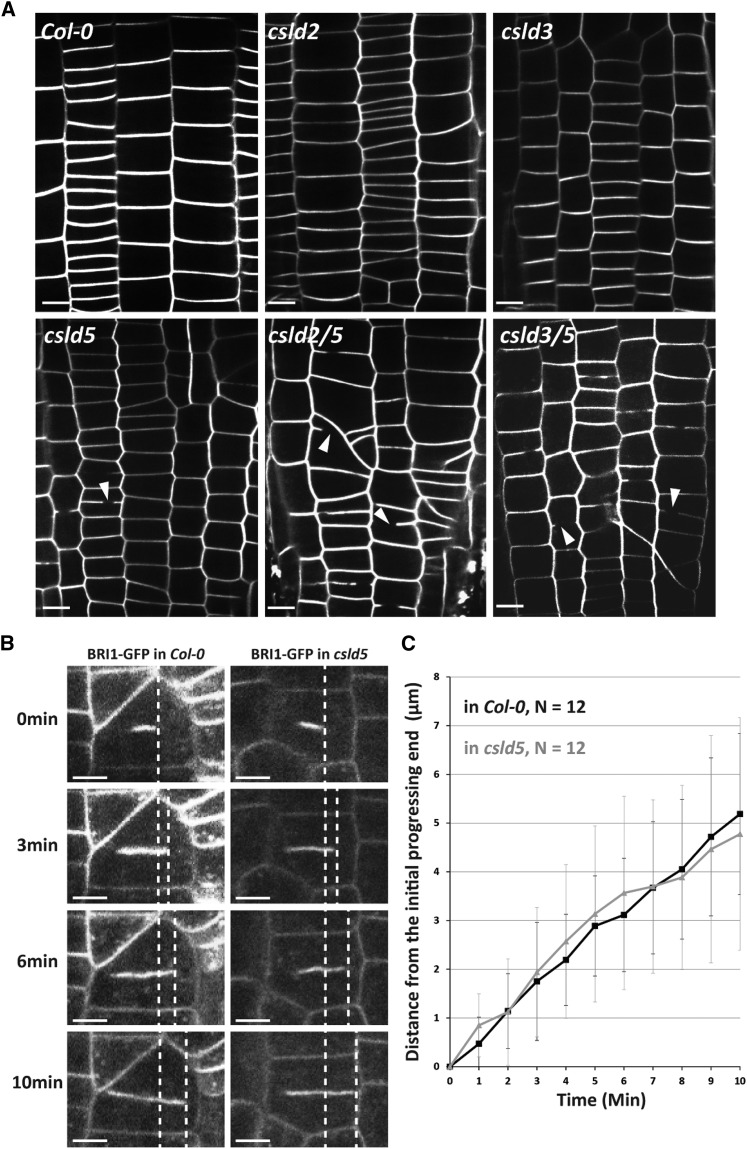
In Arabidopsis, CSLD2, CSLD3, and CSLD5 are expressed in a variety of cells and tissues, and while both csld2 and csld3 mutants have been previously shown to display root hair development phenotypes (Wang et al., 2001; Bernal et al., 2008) (Supplemental Figure 1A), they are otherwise indistinguishable from Col-0 (Supplemental Figures 1C to 1F). Loss of CSLD5 function does not appear to affect root hair development, and examination of root hair defects in csld2/5 and csld3/5 double mutants do not appear to be additive (Supplemental Figures 1A and 1B). While csld5 mutants display no root hair defects, they have measurably shorter roots, smaller rosettes, and slightly reduced overall stature when compared with Col-0 controls (Bernal et al., 2007) (Supplemental Figures 1A to 1F). As observed in developing stomatal cells, cell wall stubs were observed in root cortical cells in csld5, but not csld2 or csld3, mutants (Figure 2A, arrowheads). Both the overall plant stature phenotypes (Yin et al., 2011), and percentage of root cortical cells displaying cell wall stubs seen in csld5 mutants were dramatically enhanced in csld2/5 or csld3/5 double mutant backgrounds (Supplemental Figures 1C to 1F; Figure 2A).
Figure 2.
CSLD5 Defects in Cell Division Are Enhanced in csld2/5 and csld3/5 Mutants.
(A) Confocal images of root epidermal cells from 5-d-old seedlings, stained with FM4-64, showing division defects (arrowheads) in csld5 and the double mutants csld2/5 and csld3/5.
(B) Cell plate expansion rate was measured in Col-0 and csld5. BRI1-GFP was used as a cell plate marker. Dashed lines indicate the edges of elongating cell plates.
(C) Quantitative analysis did not show significant difference between the cell plate expansion rate of Col-0 and csld5.
Bars = 20 µm in (A) and 10 µm in (B). Error bars in (C) represent sd.
In plants, development and expansion of mature tissues and organs are dependent on both cell division and cell expansion (Cosgrove, 2005). As a result, the reduced stature observed in csld5, csld2/5, and csld3/5 mutants might result due primarily to defects in cell division, cell expansion, or a combination of both these processes. To investigate this, we examined the effects of csld mutations on cell morphology of leaf epidermal cells and root cortical cells (Supplemental Figure 2). We measured the overall size of mature root cortical cells from 5-d-old csld5, csld2/5, and csld3/5 mutants and Col-0 controls (Supplemental Figures 2A and 2B). Statistical analysis showed that while the mature cortical cell length did not differ significantly between wild-type plants and csld5 mutants, both csld2/5 and csld3/5 double mutant plants had significantly shorter root cortical cells (Supplemental Figure 2B). This was accompanied by reduced distance between the first mature root cortical cell and the quiescent center in csld5 mutants and an enhancement of this phenotype in both csld2/5 and csld3/5 mutants (Supplemental Figure 2C). To measure relative levels of cell expansion in leaf tissues, we examined the number of cells in a defined area in cotyledons of 5-d-old seedlings (Supplemental Figures 2D and 2E). Interestingly, we observed reduced numbers of larger epidermal pavement cells in csld5 mutants, and these effects were unchanged in both csld2/5 and csld3/5 double mutants (Supplemental Figure 2E). Because leaf cells are actually larger in the absence of CSLD5, this indicates a role in cell division and not cell expansion, in contrast to CSLD2 and CSLD3, which function in both processes. The relative impact of CSLD2 and CSLD3 on cell expansion in different tissues appears variable, possibly reflecting different contributions of other cell wall synthases, such as CESA proteins during expansion of different cell types during plant development.
To understand how cell wall gaps might be formed upon the disruption of CSLD5, we examined actively dividing cells in wild-type Col-0 and csld5 mutant plants (Figure 2B). In dividing plant cells, new cell walls are formed through the continued delivery and fusion of cell plate-forming vesicles until fusion with the limiting plasma membrane physically separates the two daughter cells (Jürgens, 2005). The incomplete cell walls observed in csld5, csld2/5, and csld3/5 mutants might be caused by impaired growth of the newly forming cell plate during cytokinesis in dividing plant cells. Alternatively, in csld mutants, this newly forming membrane compartment may not accumulate sufficient cell wall material, thereby affecting the structural integrity, which could lead to the formation of gaps in the walls between newly divided cells. To examine whether incomplete cell wall defects observed in csld5 mutants were due to impaired delivery and fusion of cell plate-forming vesicles, we utilized a membrane-localized BRI1-GFP to monitor the rate at which membranes accumulated at the cell division plane (Friedrichsen et al., 2000). Due to the low penetrance of cell wall division defects in csld5 mutants (∼1 to 3%), it was not possible to obtain movies of actively dividing cells that resulted in a cell division defect. However, we found that cell plates in csld5 mutants had an elongation velocity of 0.47 ± 0.24 µm/min, which did not significantly differ from that of Col-0, whose elongation velocity was 0.51 ± 0.14 µm/min (Figure 2B; Supplemental Movies 2 and 3; Student’s t test P value = 0.692). We interpret this to indicate that loss of CSLD5 does not impact the rate at which membrane-bounded cell plate structures are formed. Instead, loss of CSLD5 activity may impair the structural integrity of these structures, which could result in the formation of gaps in the dividing cell wall during cytokinesis.
The Dynamics of CSLD5 Proteins in Dividing Cells Is Uniquely Regulated by the Cell Cycle
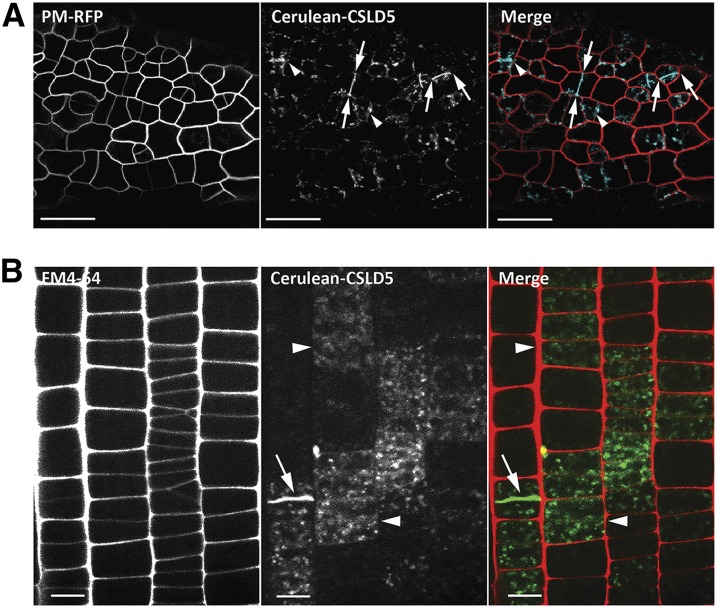
If loss of CSLD activity impairs cell wall deposition during cell division in csld5, csld2/5, and csld3/5 mutants, these proteins should be present on forming cell plates. We therefore examined whether CSLD2, CSLD3, and CSLD5 were localized to forming cell plates during cytokinesis. Fluorescent protein fusions to the N-terminal ends of full-length CSLD2, CSLD3, and CSLD5 were generated and expressed under the control of their respective endogenous promoters. These fluorescent fusions were functional and could quantitatively rescue csld2 (Supplemental Figures 3A and 3B), csld3 (Park et al., 2011), and csld5 mutant phenotypes, respectively (Supplemental Figures 3C and 3D). Both leaf epidermal cells and root cortical cells of 5-d-old seedlings expressing these fusion proteins were examined using confocal microscopy. As expected, we found that Citrine-CSLD2, eYFP-CSLD3, and Cerulean-CSLD5 were all highly enriched on cell plates in dividing cells (Figures 3A and 3B; Supplemental Figure 4). While both Citrine-CSLD2 and eYFP-CSLD3 accumulated in punctate structures in all nondividing cells in the root and aerial tissues we examined (Supplemental Figure 4), Cerulean-CSLD5 fluorescence was associated with patches of cells that were undergoing cell division (Figures 3A and 3B).
Figure 3.
Cerulean-CSLD5 Is Enriched in Cell Plates of Dividing Cells.
Distribution of Cerulean-CSLD5 in leaf epidermal cells (A) and root epidermal cells (B) of 5-d-old Col-0 seedlings. Cerulean-CSLD5 (cyan in [A]; green in [B]) localized to newly forming cell plates in dividing cells (arrows). Cell outlines were marked with either PM-RFP (mCherry-RCl2A) (A) or FM4-64 in (B); Cerulean-CSLD5 was localized to punctate subcellular compartments in some of the interphase cells that may have recently completed cell divisions (arrowheads). Bars = 25 μm in (A) and 20 μm in (B).
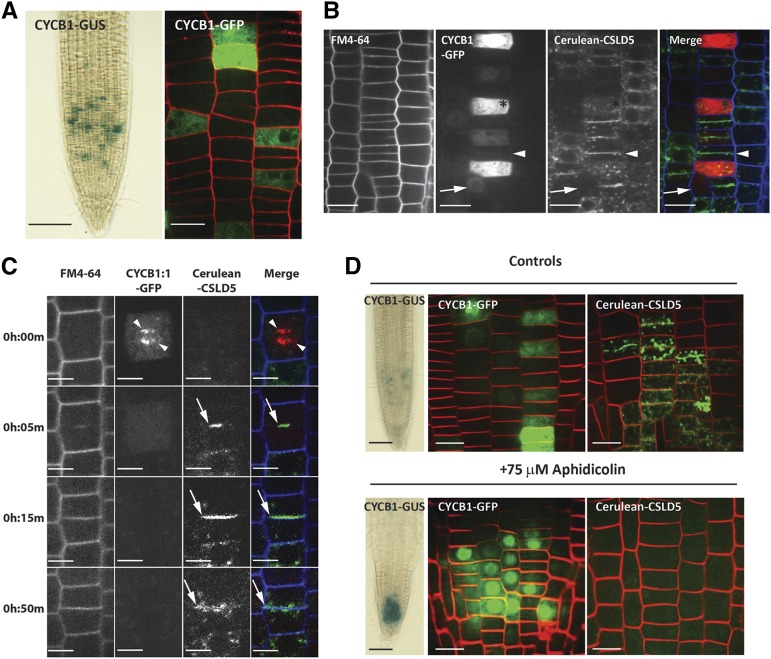
To test the idea that CSLD5 selectively accumulated during cell division, we compared the distribution pattern of Cerulean-CSLD5 and CYCB1:1, a cyclin expressed initially during S-phase, but which becomes enriched during the G2/M transition before being rapidly degraded at the metaphase-to-anaphase transition during mitosis (Colón-Carmona et al., 1999). We found that CYCB1:1 also accumulated in patches of cells that were distributed nonuniformly within the root tissues examined, similar to the Cerulean-CSLD5 accumulation pattern observed (Figure 4A). To investigate whether CSLD5 also accumulated in dividing cells during the G2/M transition as observed with CYCB1:1, we crossed Cerulean-CSLD5 into plants expressing CYCB1:1-GFP and examined the extent of overlap between these two fluorescent proteins. Surprisingly, the distribution of Cerulean-CSLD5 and CYCB1:1-GFP was nearly mutually exclusive (Figure 4B). To further examine the accumulation of Cerulean-CSLD5 and CYCB1:1-GFP during cell division, we examined the dynamics of these two proteins in time-lapse movies of dividing cells. We found that significant levels of Cerulean-CSLD5 fluorescence were observed only after the separation of CYCB1:1-GFP labeled chromatin and initiation of cytokinesis in these mitotic cells (Figure 4C; Supplemental Movie 4). Interestingly, Cerulean-CSLD5 fluorescence was rapidly lost once the newly formed cell plate had separated the two daughter cells (Figure 4C, 50 min time point), perhaps indicating that CSLD5 is rapidly downregulated upon exit of mitosis. To further test whether expression and accumulation of Cerulean-CSLD5 and CYCB1:1-GFP were linked to the cell cycle, the accumulation of these proteins was observed upon growth in media containing aphidicolin, a DNA polymerase inhibitor that locks cells in the S-phase of the cell cycle (Menges and Murray, 2002). After 2 d of growth in the presence of aphidicolin, increased numbers of root cells accumulated CYCB1:1-GFP, consistent with a blockade in the S-phase of the cell cycle (Figure 4D). On the other hand, the level of Cerulean-CSLD5 was undetectable (Figure 4D), consistent with our earlier results that Cerulean-CSLD5 did not accumulate until after loss of CYCB1:1-GFP signal during the metaphase-anaphase transition of mitosis.
Figure 4.
CSLD5 Accumulation Is Regulated by the Cell Cycle.
(A) Representative images of the distribution of a CYCB1-GUS translational fusion (left panel) and CYCB1-GFP (right panel) in root meristematic zones (green = CYCB1-GFP; red = FM4-64).
(B) The distribution of CYCB1-GFP (red) and Cerulean-CSLD5 (green) in 5-d-old seedlings stained with FM4-64 (blue). Cerulean-CSLD5 was largely absent from cells containing significant levels of CYCB1-GFP (arrows). CYCB1-GFP was absent from the cells with Cerulean-CSLD5 fluorescence on cell plates (arrowheads).
(C) CYCB1-GFP and Cerulean-CSLD5 dynamics during mitosis. Five-day-old seedlings expressing CYCB1-GFP and Cerulean-CSLD5 were examined by time lapse video microscopy. Cell peripheries of root epidermal cells were labeled by a 5-min pulse of FM4-64 (blue). CYCB1-GFP (red) initially labeled chromatids at the metaphase-anaphase transition (arrowheads; 0 h:00 m time point), but CYCB1-GFP signal was rapidly lost upon transition to cytokinesis (0 h:05 m time point). Only after CYCB1-GFP signal disappeared was Cerulean-CSLD5 observed in newly forming cell plates (arrow, 0 h:05 m and 0 h:15 m time points). Upon completion of cytokinesis, Cerulean-CSLD5 signal was depleted from the newly formed dividing cell wall and appeared in punctate cytoplasmic compartments (arrow; 0 h:50 m time point).
(D) Cerulean-CSLD5 does not accumulate in early S-phase. Treatment with 75 µM aphidicolin for 48 h locked the cell cycle in early S-phase, resulting in proliferation of cells containing CYCB1-GUS (left panels) or CYCB1-GFP (middle panels), but no Cerulean-CSLD5 signal could be detected in root epidermal cells treated with aphidicolin (right panels).
Bars = 1 mm in (A) and (D) for CYCB1-GUS, 20 μm in (A), (B), and (D) for CYCB1-GFP, and 20 μm in (B), 10 μm in (C), and 20 μm in (B) and (D) for Cerulean-CSLD5.
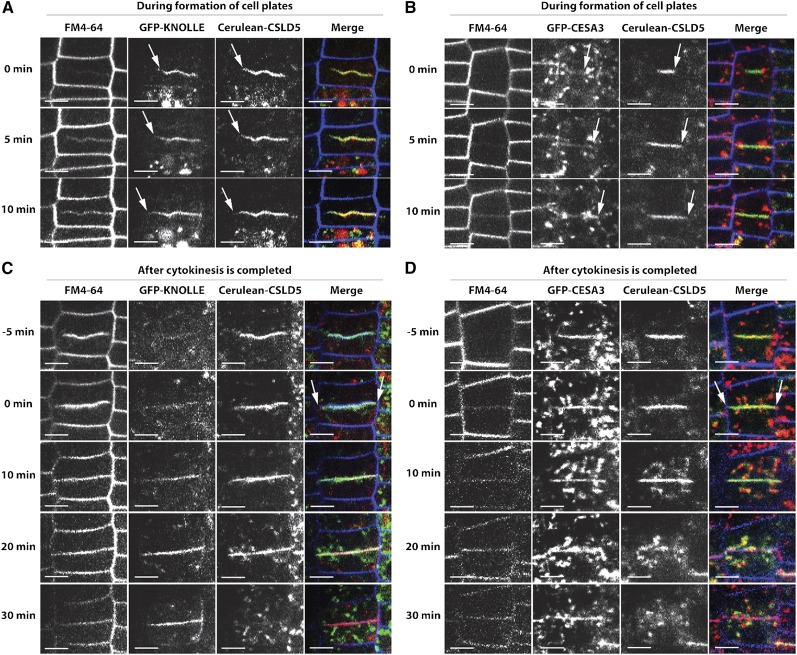
To obtain a more precise temporal-spatial analysis of the distribution of Cerulean-CSLD5 during cytokinesis, we further compared its cell plate accumulation pattern with two other cell plate-localized proteins, the cytokinesis-specific syntaxin KNOLLE (Lauber et al., 1997; Touihri et al., 2011) and the cellulose synthase CESA3 (Desprez et al., 2007; Miart et al., 2014). Cerulean-CSLD5 was crossed into lines stably expressing GFP-KNOLLE and GFP-CESA3, and cellular dynamics of these proteins were compared using time-lapse confocal microscopy. Current models of cell plate formation during cell division describe a series of successive stages, with initial targeting and fusion of Golgi-derived vesicles giving rise to tubule-vesicular networks, and these membrane networks subsequently being filled with new cell wall material to form planar fenestrated membrane sheet-like structures that extend centripetally and fuse with the plasma membranes to complete cellular division (Samuels et al., 1995; Otegui and Mastronarde, 2001; Drakakaki, 2015). In an attempt to discriminate between potential early and late roles for CSLD5, KNOLLE, and CESA3 proteins in these cell plate formation events, we segregated early cell division events (in which the forming cell plate was less than 50% of the width of the cell) from later stages of cell plate formation (in which the forming cell plate was more than 50% of the width of the cell). We found that both GFP-KNOLLE (Figure 5A; Supplemental Movie 5) and GFP-CESA3 (Figure 5B; Supplemental Movie 6) colocalized with Cerulean-CSLD5 on newly forming cell plates during cell division, and all three fluorescent fusion proteins appeared to be uniformly distributed along the length of these forming cell plates (Supplemental Figure 5A). However, while both GFP-KNOLLE and Cerulean-CSLD5 localized to newly forming cell plates at early stages during cell plate formation (Figure 5A), the subcellular distribution of GFP-CESA3 to newly forming cell plates was qualitatively distinct, and significant levels of GFP-CESA3 were maintained in punctate subcellular structures throughout cytokinesis (compare Figures 5A and 5B). To quantify these distinct subcellular dynamics, we assessed the relative fluorescence enrichment of GFP-KNOLLE, GFP-CESA3, and Cerulean-CSLD5 on forming cell plates during cytokinesis (Supplemental Figure 5B). Virtually all GFP-KNOLLE and Cerulean-CSLD5 fluorescence rapidly associated with newly forming cell plates (Figure 5A; Supplemental Figure 5C), while GFP-CESA3 remained predominantly associated with a population of punctate subcellular compartments during early stages of cytokinesis (Figure 5B; Supplemental Figure 5C). While the levels of GFP-CESA3 enrichment on cell plates gradually increased during cytokinesis progression (Supplemental Figure 5C), a significant fraction of GFP-CESA3 always remained associated with these punctate structures at all stages of cytokinesis (Figure 5B; Supplemental Figure 5C). These results are consistent with a model in which both CESA3 and CSLD5 function cooperatively to synthesize nascent cell walls during cytokinesis, but in which CSLD5 action predominates during the early stages of cell wall formation. Loss of CSLD activity in csld5 mutants would impair cell wall deposition during these early stages, leading to defects in the integrity of these newly forming cell walls.
Figure 5.
Spatio-Temporal Analysis of Cerulean-CSLD5 in Cell Plates.
Cerulean-CSLD5 (green), GFP-KNOLLE (red; [A] and [C]), and GFP-CESA3 (red; [B] and [D]) subcellular distributions were examined in dividing root epidermal cells of 5-d-old Col-0 seedlings. Representative images of colocalization in cells with newly forming cell plates ([A] and [B]; arrows) and in cells once the cell plate had contacted the plasma membrane ([C] and [D]; arrows). GFP-KNOLLE and Cerulean-CSLD5 colocalized on cell plates at both early and late stages of cytokinesis ([A] and [C]), but upon completion of cell division Cerulean-CSLD5 was lost from newly formed diving cell walls, while GFP-KNOLLE was not ([C]; compare 20 and 30 min time points). At early stages of cytokinesis, GFP-CESA3 only weakly labeled newly forming cell plates containing significant Cerulean-CSLD5 signal ([B]; arrows), but GFP-CESA3 labeling of cell plates and newly formed dividing cell walls was more pronounced (compared with [B] and [D]). Also, unlike either Cerulean-CSLD5 or GFP-KNOLLE, significant GFP-CESA3 signal was observed in punctate cytoplasmic structures at both early and late stages during cytokinesis ([B] and [D]). Bars = 10 µm.
Interestingly, analysis of time-lapse images during and after completion of cytokinesis (Figures 5C and 5D, arrows; Supplemental Movies 7 and 8) also uncovered differences between GFP-KNOLLE, GFP-CESA3, and Cerulean-CSLD5 subcellular dynamics. Upon completion of cytokinesis, individual structures containing Cerulean-CSLD5 appeared to bud off the recently formed cell plate (Figures 5C and 5D). The emergence of these structures coincided with a rapid reduction of Cerulean-CSLD5 fluorescence on membranes associated with the newly synthesized cell wall separating the two newly formed daughter cells (Figures 5C and 5D). These differences were also reflected by quantitative analysis of the relative fluorescence enrichment characteristics of Cerulean-CSLD5, which was reduced by ∼50% within 10 min of the apparent completion of cytokinesis (Supplemental Figure 5C). By contrast, the fluorescence level of GFP-KNOLLE on newly formed cell walls remained high and actually increased slightly during the 30 min after completion of cytokinesis (Figure 5C). KNOLLE has also been shown to be removed from cell plates and degraded in vacuolar compartments upon completion of cytokinesis (da Silva Conceição et al., 1997). The distinct subcellular dynamics observed for GFP-KNOLLE and Cerulean-CSLD5 upon completion of cytokinesis may indicate distinct mechanisms of removal and turnover of these proteins. Fluorescence levels of GFP-CESA3 remained largely unchanged during the 30-min period after completion of cytokinesis (Figure 5D; Supplemental Figure 5C). Based on these observations, we propose three distinct stages during cell plate formation in plant cells undergoing cytokinesis. During initiation, we observed significant labeling of cell plates by both Cerulean-CSLD5 and GFP-KNOLLE (Figure 5A). This labeling was accompanied by a distinct lack of these proteins in punctate subcellular structures. By contrast, at these early stages of cell division, GFP-CESA3 was primarily localized in punctate subcellular structures, and little was detected on nascent cell plate structures (compare Figures 5A and 5B, zero time points). At later stages, both Cerulean-CSLD5 and GFP-KNOLLE were maintained on growing cell plates, and GFP-CESA3 displayed increasing levels of recruitment to these structures, perhaps reflecting the reinforcement of these newly forming cell walls (Figure 5B, 5 and 10 min time points; Supplemental Figure 5C). Completion of cytokinesis was defined as contact and fusion of the expanding cell plate to the plasma membrane. At this point, Cerulean-CSLD5 fluorescence was rapidly lost from membranes associated with the newly formed dividing cell wall, while GFP-CESA3 levels on these membranes remained largely unchanged and GFP-KNOLLE association transiently increased (Figures 5C and 5D). The distinct dynamics of Cerulean-CSLD5 observed during these three stages of cytokinesis highlight the specific targeting of this protein during plant cytokinesis and support the possibility that CSLD5 is uniquely regulated during this stage of the plant cell cycle.
CSLD5 Proteins Are Rapidly Destabilized upon the Completion of Cytokinesis
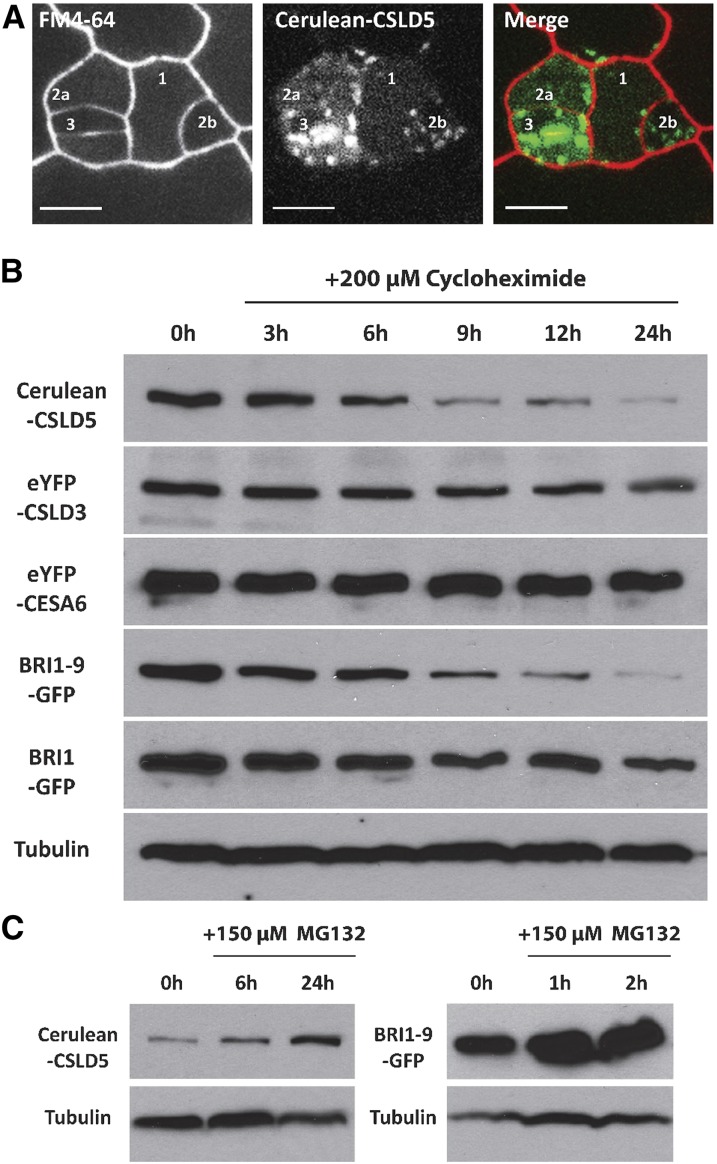
To test if the rapid depletion of Cerulean-CSLD5 fluorescence in cells that had completed cytokinesis might reflect an active removal of these proteins, we reexamined the distribution of Cerulean-CSLD5 in self-renewing meristemoid cells. Cerulean-CSLD5 fluorescence intensity was highest in cells that appeared to have just finished cell division, with successively reduced levels of fluorescence observed in cells that had undergone earlier divisions (Figure 6A; Supplemental Figure 6). Consistent with this, time-lapse analysis of stomatal cell divisions also showed that upon completion of cytokinesis, Cerulean-CSLD5 fluorescence levels were dramatically reduced (Supplemental Movie 9).
Figure 6.
Analysis of the Stability of Cerulean-CSLD5.
(A) Persistence of Cerulean-CSLD5 fluorescence is negatively associated with cell age. In stomatal lineages, where birth order can be surmised by position and morphology, most recently divided cells (guard cells, #3) contain highest CSLD5 fluorescence levels, the second newest cells (#2a and #2b) have intermediate levels of CSLD5, and nondividing SLGCs (#1) have lowest levels of CSLD5.
(B) Protein turnover rates of CSLD and other cellular proteins treated with 200 µM cycloheximide. Total proteins were extracted from 5-d-old seedlings at each time point and the relative levels were determined using immunoblotting with anti-GFP antibodies. Cerulean-CSLD5 levels rapidly decreased during the time course, while levels eYFP-CSLD3 and eYFP-CESA6, two structurally similar proteins, were not significantly reduced. A destabilized mutant of BRI1, BRI1-9-GFP, also displayed rapid protein turnover kinetics, while wild-type BRI1-GFP levels were stable. Tubulin was used as a loading control.
(C) Stabilization of Cerulean-CSLD5 turnover by treatment with MG132. Five-day-old seedlings were grown in media containing 150 µM MG132 for 24 h. The level of Cerulean-CSLD5 was increased after 6 h (left panels). BRI1-9-GFP, which is turned over in a ubiquitin-dependent manner, was also stabilized under these conditions (right panels). Tubulin was used as a loading control.
To confirm Cerulean-CSLD5 is an unstable protein, we blocked new protein synthesis by treating 5-d-old Arabidopsis seedlings with cycloheximide and then compared protein turnover rates of several membrane proteins. Overall accumulation of Cerulean-CSLD5 was reduced significantly during the time course (Figure 6B) but was not observed for either eYFP-CSLD3 or eYFP-CESA6 (Figure 6B).
Rapid Cerulean-CSLD5 turnover might be due to posttranslational ubiquitin modification (Marrocco et al., 2010). We were curious whether CSLD5 turnover might also be regulated by ubiquitin-dependent processes. To test this, we treated seedlings with the 26S proteasome inhibitor, MG132, and examined the effects on protein accumulation characteristics of these two proteins. As with BRI-1-9-GFP, a protein known to be turned over in a ubiquitin-dependent process (Hong et al., 2009) (Figure 6B), treatment with MG132 stabilized Cerulean-CSLD5 proteins, albeit with somewhat slower kinetics (Figure 6C). This indicated that the rapid loss of Cerulean-CSLD5 signal observed upon completion of cytokinesis likely involves ubiquitin-dependent steps that regulate its stability.
CSLD Proteins Are Ubiquitinated and Interact with APC Components
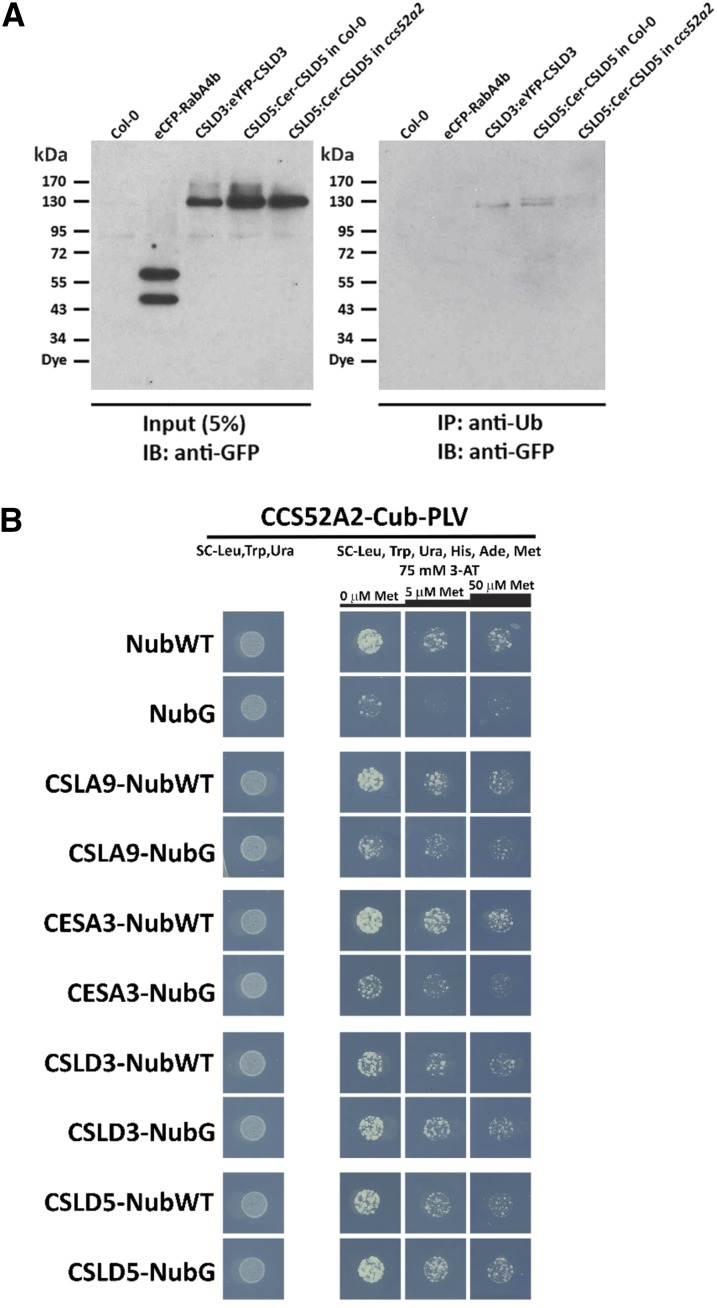
If CSLD5 proteins are actively destabilized in a ubiquitin-dependent fashion, we might observe ubiquitin-modified forms of these proteins. To test this, we first used antiubiquitin (anti-Ub) antibodies to immunoprecipitate ubiquitin-modified proteins from plants expressing eYFP-CSLD3, or Cerulean-CSLD5, under control of their endogenous promoters (Figure 7A). As negative controls, we used plants expressing eCFP-RabA4b or untransformed wild-type plants. Relative amounts of FP-tagged proteins present in detergent-solubilized membrane fractions were assessed by immunoblotting with anti-GFP antibodies (Figure 7A, left panel). While no enrichment of ubiquitin-modified proteins was detected in Col-0 and eCFP-RabA4b immunoprecipitated fractions, a single eFYP-CSLD3 band was detected at ∼130 kD (Figure 7A, right panel). Interestingly, while eYFP-CSLD3 was immunoprecipitated as a singlet of ∼130 kD, Cerulean-CSLD5 expressed behind its endogenous CSLD5 promoter (CSLD5pro:Cer-CSLD5) was detected in anti-Ub immunoprecipitated fractions as a doublet (Figure 7A, right panel). These results suggest that both CSLD3 and CSLD5 proteins can be posttranslationally modified with ubiquitin but that the specific nature of these ubiquitin modifications may be distinct.
Figure 7.
CSLD Proteins Are Ubiquitin-Modified and Interact with the APC Activator Protein CCS52A2.
(A) Immunoprecipitation of ubiquitin-modified Arabidopsis proteins. Total proteins isolated from 5-d-old seedlings expressing eCFP-RabA4b, EYFP-CSLD3, or Cerulean-CSLD5 either in wild-type (Col-0) or ccs52a2 mutant backgrounds were subjected to immunoprecipitation with anti-Ub antibodies. Total proteins equivalent to 5% of input or antiubiquitin (anti-Ub)-immunoprecipitated proteins were separated by SDS-PAGE and fluorescently tagged proteins were detected by immunoblotting with anti-GFP antibodies.
(B) Detection of CCS52A2-CSLD interaction by split-ubiquitin yeast two-hybrid assay. Diploid yeast coexpressing CCS52A2-Cub-PLV bait constructs and various NubWT/NubG prey constructs. CCS52A2 was fused with Cub-PLV tag driven by a Met-repressible promoter. CSLA9, CESA3, CSLD3, and CSLD5 proteins were C-terminal tagged with both NubWT and NubG, respectively. Yeast were grown on vector-selective plates (SC-Leu, -Trp, -Ura) and interaction-selective plates (75 mM 3-AT, SC-Leu, -Trp, -Ura, -Ade, -His, -Met) with increasing Met concentrations (0, 5, and 50 µM Met). Yeast growth was recorded after 20 h for the vector-selective plates and 60 h for interaction-selective plates.
In eukaryotic cells, cell cycle progression and ubiquitin-mediated protein turnover are linked by the APC, a multisubunit E3 ubiquitin ligase (Marrocco et al., 2010). The APC regulates cell cycle progression by degrading downstream targets, such as CYCB1:1 (Zheng et al., 2011). The substrate specificity of the APC differs at different stages of the cell cycle, and this is regulated by association of the APC with distinct activator proteins such as CDC20 and CDH1 (Heyman and De Veylder, 2012). In Arabidopsis, eight distinct APC activators have been described, five CDC20 genes and three plant CDH1 homologs, called Cell Cycle Switch Protein 52 A1 (CCS52A1), (CCS52A2), and B (CCS52B) (Fülöp et al., 2005). Of these activator proteins, only CCS52A1 and CCS52A2 appear to function during late M and G1 phases of the cell cycle, while CDC20 proteins and CCS52B are expressed during the G2/M transition and in early stages of mitosis (Fülöp et al., 2005). Since the proteolytic turnover of Cerulean-CSLD5 appeared to occur after completion of cytokinesis, we hypothesized that CCS52A1 or CCS52A2 genes may impact the stability of Cerulean-CSLD5. To test this possibility, we examined whether the APC-activator, CCS52A2, could interact with CSLD proteins using a mating-based, split-ubiquitin yeast two-hybrid system specifically designed to detect interactions between integral membrane proteins (Obrdlik et al., 2004). Full-length CCS52A2 was attached to the N terminus of a construct consisting of the C-terminal ubiquitin fragment transcription factor fusion protein (CCS52A2-Cub-PLV). CSLD proteins, CSLD3 and CSLD5, the cellulose synthase CESA3, and the mannan synthase CSLA9, were tested for interaction with CCS52A2 by attaching their coding sequences to N-terminal fragments of either wild-type ubiquitin (NubWT) or a mutated version (NubG) that only interacts with Cub fusions upon association of CCS52A2 with the tested fusion protein. Yeast expressing NubWT and Cub fusions all grew in selection conditions, indicating that both Nub and Cub fusion constructs accumulated in yeast membranes (Figure 7B, NubWT lanes). However, significant growth was observed in yeast expressing CSLD3-NubG and CSLD5-NubG constructs, but not for those expressing CSLA9-NubG, CESA3-NubG, or NubG alone (Figure 7B, NubG lanes). In all cases, growth was inhibited in the presence of methionine, indicating the presence of both Nub and Cub fusion constructs was required for growth (Figure 7B, compare 0 µM Met, 5 µM Met, and 50 µM Met). These results are consistent with direct interaction between CCS52A2 and CSLD proteins.
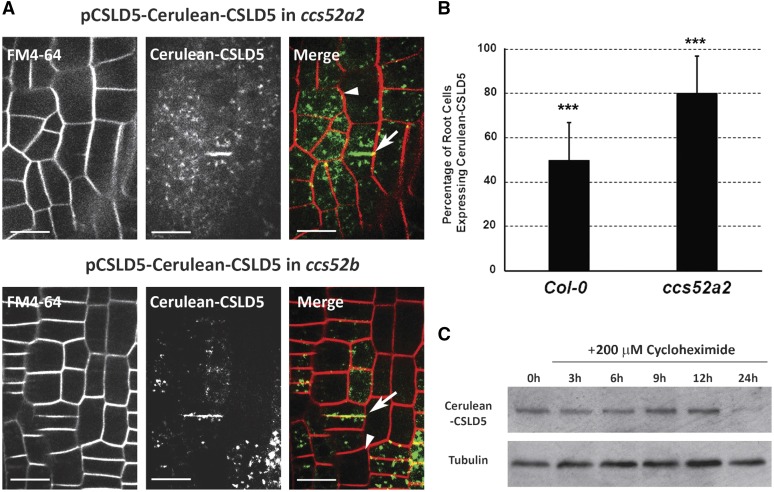
To further examine the role of CCS52A2 mutation on CSLD5 accumulation during the cell cycle, we transformed Cerulean-CSLD5 under the control of its own promoter into different ccs52 knockout lines. Disruption of ccs52a1 appeared to be lethal, since we were unable to obtain homozygous T-DNA insertional mutants. However, we were able to obtain homozygous ccs52a2 mutants, although these mutants displayed significant growth defects that resulted in a severe dwarf phenotype. Interestingly, the association between Cerulean-CSLD5 accumulation and cell division (Figure 3A) was no longer observed in ccs52a2 mutants, and instead Cerulean-CSLD5 fluorescence in this mutant background resembled the uniform accumulation patterns observed for both Citrine-CSLD2 and eYFP-CSLD3 proteins in root epidermal cells (Figure 8A). To quantify this altered accumulation pattern, the percentage of cells in root meristematic zones containing Cerulean-CSLD5-labeled punctate structures was quantitated for 38 Col-0 and ccs52a2 mutant seedlings, respectively. In the wild-type background, 49.8% of root cells displayed discernable accumulation of Cerulean-CSLD5, whereas in the ccs52a2 mutant background, this percentage was significantly increased to 80.1% (Figure 8B). This loss of regulated accumulation of Cerulean-CSLD5 appeared to be specific to loss of CCS52A2 gene function, as the cell division-associated accumulation pattern of Cerulean-CSLD5 was unchanged in the ccs52b mutant background (Figure 8A). Further supporting the role of CCS52A2 in cell cycle-dependent regulation of Cerulean-CSLD5 stability, we observed that the rapid Cerulean-CSLD5 protein turnover kinetics observed in wild-type Col-0 plants was lost in the ccs52a2 mutant background (Figure 8C), and dramatically reduced levels of Ub-modified Cer-CSLD5 were detected in anti-Ub immunoprecipitations from ccs52a2 mutant plants expressing Cer-CSLD5 (Figure 7A; compare CSLD5:Cerulean-CSLD5 in Col-0 and CSLD5:Cerulean-CSLD5 in ccs52a2 lanes). Based on these results, we concluded that the turnover of CSLD5 might be directly, or indirectly, mediated via APCCCS52A2-regulated processes. These results indicate that the regulated accumulation CSLD5 protein during mitosis, and its active removal upon completion of cytokinesis, appears to result from the participation of core elements of the cell cycle regulatory machinery.
Figure 8.
Cerulean-CSLD5 Turnover Characteristics Are Altered in ccs52a2 Mutants.
(A) Cerulean-CSLD5 fluorescence accumulated broadly in both dividing root epidermal cells (arrowhead) and in nondividing cells (arrows) in a ccs52a2 mutant background (upper panels), but selective Cerulean-CSLD5 fluorescence remained associated with dividing cells (arrowhead) in a ccs52b mutant background (lower panels).
(B) Confocal images were obtained from FM4-64 stained 5-d-old seedlings expressing Cerulean-CSLD5 in either the wild-type Col-0 or ccs52a2 mutant background (n = 17 seedlings; 1597 and 1451 cells in wild-type Col-0 and ccs52a2 mutant backgrounds, respectively). The proportion of cells which express Cerulean-CSLD5 is presented as a percentage of the total number of nondividing cells within each field. A statistically significant difference was detected in the percentage of non-dividing cells in the ccs52a2 mutant background compared with Col-0; ***P < 0.001 as determined by the Mann-Whitney rank sum test. Error bars represent sd.
(C) Cerulean-CSLD5 proteins were stabilized in the ccs52a2 mutant background. Seven-day-old seedlings were treated with 200 µM cycloheximide and total proteins were extracted at each time point listed. The level of Cerulean-CSLD5 remained stable 12 h after the treatment. Tubulin was used as a loading control.
DISCUSSION
Plant cells, which are encased within load-bearing cell walls, perform cytokinesis by synthesizing new cell walls to separate daughter cells. In Arabidopsis, CSLD2, CSLD3, and CSLD5 are all expressed and play important roles in a variety of vegetative tissues during plant growth and development. Examination of the subcellular characteristics of functional, fluorescent fusions of these CSLD proteins revealed that all three are targeted to newly forming cell plates in dividing cells (Figures 3 and 5A; Supplemental Figure 4) and likely all contribute to the synthesis of dividing cell walls during cytokinesis. However, among plants bearing single mutations in CSLDs, aberrant stomatal development and cell division defects were only observed in plants containing the csld5 mutant allele. We interpret this as evidence of a unique function for CSLD5 during cell division, which we believe is associated with cell cycle-regulated control of the activity of CSLD5 (Figure 9). Unlike CSLD2 and CSLD3 proteins, CSLD5 accumulation was tightly associated with cells undergoing cell division. Furthermore, cells arrested in the S phase of the cell cycle were depleted of CSLD5, indicating it may be preferentially expressed in G2/M phases (Figures 3, 4, and 9B). Consistent with this, putative binding sites for MYB3R1 and MYB3R4, two transcription factors known to regulate the G2/M phase-specific genes such as KNOLLE (Haga et al., 2011), are found in the promoter sequences of CSLD5, but not CSLD2 or CSLD3. Upon completion of cytokinesis, CSLD5 protein was rapidly depleted from the cells, and this was associated with the observation that CSLD5 was much less stable than other CSLD proteins and related CESA cellulose synthases (Figure 6). CSLD5 protein turnover appears to be regulated by ubiquitin-mediated processes, as the 26S proteasome inhibitor, MG132, stabilizes this protein, and this protein is immunoprecipitated by antiubiquitin antibodies (Figure 7A). Regulation of CSLD5 function can be further linked to the cell cycle through interaction with the APC activator CCS52A2 (Figure 7B), and CSLD5 is stabilized in ccs52a2 mutants that compromise the E3-ubiquitin ligase activity of APCCCS52A2 cell cycle regulatory complexes (Figures 8 and 9B).
Figure 9.
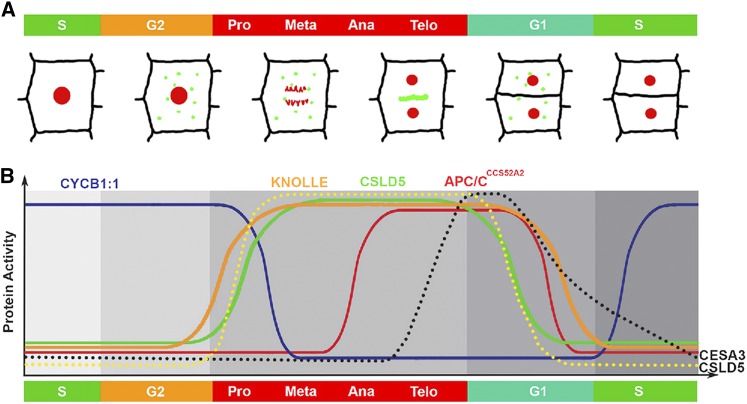
A Model for Cell Cycle-Regulated CSLD Functions during Cell Division.
(A) During interphase (G1, S, and G2 phases), CSLD2 and CSLD3 localize to punctate subcellular structures in the cytoplasm, while CSLD5 accumulation is suppressed (B). Upon entry into mitosis, expression of CSLD5 ([B], solid green line) and the cytokinesis-specific syntaxin, KNOLLE ([B], solid orange line) is upregulated. Once completion of the metaphase-anaphase checkpoint is accomplished as monitored by proteolytic turnover of CYCB1:1 ([B], solid blue line), CSLD5 ([B], dotted yellow line) as well as CSLD2 and CSLD3 are selectively targeted to newly forming cell plates (A) where these proteins synthesize polysaccharides of the new cell plate. While both KNOLLE and CSLD proteins are continuously present during cell plate synthesis, significant levels of CESA3 cellulose synthases are observed only at later stages of cell plate synthesis ([B], dotted black line). Upon completion of cytokinesis and passage of the M/G1 checkpoint, APCCCS52A2 E3-ubiquitin ligase activity ([B], solid red line) regulates the removal and proteolytic turnover of CSLD5 proteins.
Based on our observations, CSLD5 appears to be required for all plant cell divisions. This would be consistent with earlier observations that CSLD5 is most highly expressed in the shoot apical regions of Arabidopsis (Bernal et al., 2007). Furthermore, the putative rice (Oryza sativa) ortholog of Arabidopsis CSLD5, Os-CSLD4, was shown to be expressed in M-phase cells, and CSLD5 expression was associated with dividing cells in Arabidopsis shoot apical meristems (Yoshikawa et al., 2013; Yang et al., 2016). It is interesting that this gene was the sole member of the closely related CSLD and CESA cell wall synthase gene families to be enriched in the asymmetrically dividing cell population of the early stomatal lineage (Adrian et al., 2015) (Figure 1B). In addition, while csld5 mutants display cell division defects in multiple root and aerial tissues, loss of CSLD5 function results in much more severe stomatal developmental defects than observed in other tissues. This may reflect an increased requirement for CSLD5 function during the asymmetric and symmetric cell divisions that occur in rapid succession during stomatal cell division in Arabidopsis. This increased cell type-specific expression is likely provided by superimposing SPCH transcriptional control (Lau et al., 2014) (Figure 1C) upon the cell cycle-regulated expression of the CSLD5 gene.
In plants, the CESA cellulose synthase family is contained within a larger family of putative glycosyltransferases, termed the CSL gene family (Richmond and Somerville, 2001). These CSL gene families display similarity to CESA sequences and have been proposed to function during the synthesis of distinct plant cell wall polysaccharides. Members of the CSLA family have been demonstrated to synthesize β-1,4-linked mannans and glucomannans (Liepman et al., 2005; Goubet et al., 2009), and CSLCs are implicated in the synthesis of the β-1,4-linked glucan backbone of xyloglucan (Cocuron et al., 2007). Similarly, members of the CSLF and CSLH families have been linked to β-1,3-1,4-mixed linked glucan synthesis (Burton et al., 2006; Dwivany et al., 2009). However, the biochemical activities of many of the other CSL protein families remain relatively uncharacterized. Mannan synthase activity was shown to be increased in microsomal membrane fractions isolated from plants overexpressing CSLD proteins, although these activities were not determined for purified proteins (Yin et al., 2011; Verhertbruggen et al., 2011). Intriguingly, in these studies, 35S-driven expression of CSLD5 alone was sufficient for increased mannan production, whereas 35S-driven coexpression of both CSLD2 and CSLD3 was necessary to observe increased mannan synthase activity (Yin et al., 2011; Verhertbruggen et al., 2011). However, CSLD proteins apparently undergo cell-specific changes in subcellular localization during plant growth and development. CSLD3, and possibly CSLD2 also, undergo root hair-specific redistribution to apical plasma membranes in root hair cells (Park et al., 2011). Here, we describe cell cycle-specific targeting and accumulation of CSLD2, CSLD3, and CSLD5 to newly forming cell plates of dividing cells. At present, it is unclear whether the altered accumulation and subcellular distributions of these CSLD proteins may affect their biosynthetic activities.
At early stages of cytokinesis, CSLD5 accumulates in newly forming cell plate compartments more prominently than does CESA3 (Figure 7). This might indicate that CSLD5 synthesizes a polysaccharide scaffold for later cellulose deposition. In current models of cell plate formation, callose, a β-1,3-glucan polymer, is initially deposited in newly forming cell plates and then this is later replaced by cellulose and other polysaccharides (Samuels et al., 1995; Chen and Kim, 2009; Drakakaki, 2015). Therefore, it is possible that CSLD5 synthesizes callose, which is later replaced by cellulose produced by CESA proteins. However, callose synthases that participate in cell division have previously been identified (Hong et al., 2001; Chen et al., 2009; Thiele et al., 2009; Guseman et al., 2010). While CSLD proteins do not share significant sequence similarity to callose synthases, they do share significant similarities with CESA proteins. Recent observations with cellulose-specific dyes and the carbohydrate binding module, CBM3a, which is selective for crystalline cellulose, have indicated that β-1,4-glucan polysaccharides are present at earlier stages of cell plate deposition than previously thought (Miart et al., 2014). CSLD3 was recently shown to accumulate in apical plasma membranes in root hair cells, where new cellulose deposition likely occurs. Furthermore, a CSLD3 chimera that contains a CESA6 catalytic domain can functionally rescue csld3 root hair phenotypes, indicating that CSLD3 might synthesize cellulose-like β-1,4-glucan polysaccharides in the tips of growing root hairs (Park et al., 2011). The high degree of sequence similarity, the corecruitment to newly forming cell plates, and the partial functional redundancy observed among CSLD2, CSLD3, and CSLD5 support the possibility that CSLD5 may also synthesize cellulose-like β-1,4-glucan polysaccharides. If that is the case, then CSLD5 might participate in the synthesis of a transient cellulose framework for further cellulose deposition, which could be performed by CESA proteins. It also is possible that CSLD proteins might synthesize mannan polysaccharides; however, mannan deposition has not been observed at the early stages of cell plate formation, and significant levels of mannan polysaccharides are primarily observed in thickened secondary walls in Arabidopsis (Handford et al., 2003). Clearly, further study into the nature of the polysaccharides synthesized by members of the CSLD family of proteins is required.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana lines used in this study were derived from the Col-0 background. The csld3 (kjk-2) (Favery et al., 2001) mutants were received from Liam Dolan (Oxford University). The csld2 (SALK_119808), csld5 (SAIL_724_G04), ccs52a1 (SALK_083656), ccs52a2 (SALK_073708), and ccs52b (SALK_098269) were obtained from the ABRC. The CYCB1:1-GUS line (Colón-Carmona et al., 1999) was kindly provided by John Schiefelbein (University of Michigan), and CYCB-GFP (Brownfield et al., 2009) was kindly provided by Peter Doerner (University of Edinburgh). We received eGFP-CESA3 (Desprez et al., 2007) and eYFP-CESA6 (Paredez et al., 2006) lines from Chris Somerville (University of California, Berkeley). The BRI1-GFP and BRI1-9-GFP lines (Hong et al., 2008) were kindly provided by Jianming Li (University of Michigan). The GFP-KNOLLE line (Boutté et al., 2010) was received from Gerd Jurgens (University of Tubingen). The eYFP-CSLD3 line was previously reported (Park et al., 2011). Arabidopsis seeds were surface-sterilized for 15 min in 30% sodium hypochlorite/0.02% Triton X-100 solution. The seeds were washed with sterilized double-distilled water and stored at 4°C overnight before being transferred to one-quarter-strength Murashige and Skoog (MS) growth medium. Seedlings were germinated and grown in growth chambers. After 7 to 14 d, the seedlings were transferred from plates to soil and grown in growth rooms. The seedlings/plants were grown at 21°C in long-day conditions (16 h of light/8 h of dark).
Expression of Cell Wall Genes in the Stomatal Lineage
Expression values for CESA and CSLD families in FACS-isolated stomatal lineage cells were derived from data sets associated with Adrian et al. (2015) following protocols defined in that publication. ChIP-seq traces were generated in online visualization tools from DNAnexus using data from Lau et al. (2014).
Root Hair and Root Phenotype Analysis
The root hair morphology was examined using 5-d-old seedlings. To measure the root hair length, the regions with mature root hairs were recorded using the 4× lens (fluorescent, NA = 0.13) on a Nikon Eclipse 6600 microscope. The length of mature root hairs in these images was measured using ImageJ (NIH). To record the root hair growth defects, the images of root hairs of interest were collected using the 10× lens (apochromatic, NA = 0.45). Seven-day-old seedlings were grown vertically and used for root length comparison. The plates were scanned and the length of the roots was measured using ImageJ (NIH).
Mature Root Cortical Cell Length, Cell Number in the Root Meristematic Zone, and Cell Number in the Leaf Epidermis Measurements
To measure the root cortical cell length, the seedlings were first stained by FM4-64 (Life Technologies) to visualize the edges of the cells. Mature root cortical cells were defined as the cortical cells above the first emerging root hair cell. The length of five continuous cortical cells counting from the first mature one was measured using ImageJ (NIH). To estimate the cell number in the root meristematic zone, the distance from QC to the initiation site of the elongation zone, defined as the point where the length of a cortical cell became larger than its width, was measured using ImageJ (NIH). To measure the number of leaf epidermal cells, the leaf epidermis at the distal end of the leaf was examined. A region of 300 µm × 300 µm was randomly assigned and all the nonstomatal cells in this region, including the cells flanking the edge of the region, were counted.
Confocal Microscopy
The fluorescent imaging was performed with an Olympus spinning disk confocal using a 20× dry lens (apochromatic, NA = 0.70) or a 60× oil lens (plan-apochromat, NA = 1.42). Metamorph Advanced software (Molecular Devices) was used for image capture. For multichannel imaging, the channels were switched sequentially from long wavelength to short wavelength. The signal from FM4-64 was excited at 561 nm and collected at 607 nm. The signal from YFP/Citrine was excited at 515 nm and collected at 562 nm. The GFP signal was excited at 488 nm and collected at 525 nm. The Cerulean signal was excited at 445 nm and collected at 483 nm. For time-lapse imaging, 30 s was used as the time interval unless specified. Confocal and differential interference contrast imaging of leaf epidermal cells was performed as described by Lau et al. (2014). Cell outlines were visualized by either the plasma membrane marker pML1:mCherry-RCI2A or with propidium iodide (Molecular Probes; P3566; 0.1 mg/mL). Stomatal defects were classified as either stomata in contact or cytokinesis defects. For the latter, guard cells were scored that failed to form a complete dividing wall and a pore (e.g., Figure 1D, lower right image). For quantification of stomatal phenotypes, seedlings 5 d after initiation were cleared in ethanol: acetic acid (7:1) solution and mounted with Hoyer’s solution before differential interference contrast imaging with a Leica DM2500 microscope (40×, 0.157 mm−2 field of view). FIJI was used for image processing and quantification (Schindelin et al., 2012). BASLp:YFP- BASL-expressing plants were used as described by Dong et al. (2009). Time-lapse imaging was conducted as described by Davies and Bergmann (2014).
Cell Cycle Inhibition
Five-day-old seedlings were transferred to liquid one-quarter-strength MS medium containing 75 μM aphidicolin (Sigma-Aldrich) for 48 h before the seedlings were used for GUS reporter gene analysis or for confocal microscopy.
GUS Staining
Histochemical analysis was performed on seedlings expressing CYCB1:1-GUS. Five-day-old seedlings were transferred to GUS staining solution (0.1 M NaPO4, pH 7.0, 10 mM EDTA, pH 7.0, 0.75 mM potassium ferricyanide, 0.75 mM potassium ferrocyanide, 1% Triton X-100, and 0.4 mM X-glucoronide) and incubated at 37°C for 1 h. The stained seedlings were directly used for accumulation pattern analysis.
Quantitative Fluorescence Imaging
Cell Plate Elongation Rate Measurement
BRI1-GFP was used as a membrane marker to track the elongation rate of cell plates in 5-d-old Col-0 and csld5 seedlings. The elongation rate of the cell plate edges was measured on a minute-by-minute basis using ImageJ (NIH).
Relative Fluorescence Enrichment Analysis
The relative fluorescence enrichment was used to examine the dynamics of GFP-KNOLLE, GFP-CESA3, and Cerulean-CSLD5 during cytokinesis. It was defined as the total fluorescence on the cell plate divided by the total fluorescence in the dividing cell. Elongating cell plates and total cellular areas were selected manually in ImageJ based on the presence of enriched fluorescence on the cell plate and plasma membrane-associated FM4-64 labeling, respectively. Total fluorescent signal within these selected regions was calculated using the integrated density function of Image J (NIH).
Root Cell Cerulean-CSLD5 Expression Analysis
The number of cells from root meristematic zones of 5-d-old seedlings, which express Cerulean-CSLD5 in nondividing root epidermal cells, defined as cells that lack a visible expanding cell plate, was determined. The proportion of cells that express Cerulean-CSLD5 is presented as a percentage of the total number of nondividing cells within each field. Statistical significance, P < 0.001, was determined by the Mann-Whitney rank sum test (Mann and Whitney, 1947).
Guard Cell Lineage Quantification
Representative images of stomatal guard cell lineages from propidium iodide-stained 5-d-old Col-0 seedlings expressing Cerulean-CSLD5 were assigned cell birth order and used for quantification of Cerulean-CSLD5 fluorescence. Fluorescence intensity of individual cells was determined in ImageJ (NIH). Corrected total cell fluorescence (CTCF) was calculated as previously described (Gavet and Pines, 2010). CTCF = integrated density of cerulean fluorescence intensity – (cell area in pixels * average mean background fluorescence for 3 regions of interest outside of the cells).
Split-Ubiquitin Yeast Two-Hybrid Analysis
Mating-based split-ubiquitin yeast two-hybrid methods (mbSUS) were performed essentially as previously described (Obrdlik et al., 2004). Two haploid yeast strains, THY.AP4 (MATa) and THY.AP5 (MATα), were used in the mbSUS assay (Obrdlik et al., 2004). CCS52A2, CESA3, CSLA9, CSLD3, and CSLD5 coding sequences were first inserted into the pENTR 3C Dual Selection vectors (Thermo Fischer Scientific) and transferred into the destination vectors MetYC_GW (Cub-PLV), XNWT_GW (NubWT), and XN21_GW (NubG) (https://associomics.dpb.carnegiescience.edu/Associomics/Vectors_and_Clones.html).
The CCS52A2 bait construct was then transformed into the THY.AP4 strain and selected by growth on SC-Leu media, while the prey constructs were transformed into the THY.AP5 strain and selected by growth on SC-Trp media. One milliliter of each stationary culture was pelleted and resuspended in 200 μL of YPD medium. Mating mixes for testing individual yeast two-hybrid interactions in diploid yeast were generated by mixing 15 μL each of THY.AP4 and THY.AP5 yeast suspensions. Four microliters of these mating mixes was grown on a YPD plate for 12 h at 28°C. Diploid yeast colonies were resuspended in double distilled water, adjusted to OD600 = 0.1, spotted onto SC-Leu, Trp, Ura or SC-Leu, Trp, Ura, Ade, His, Met selection plates, and incubated at 28°C. Yeast growth was recorded every 12 h.
Immunoprecipitation and Immunoblot Analysis
Total protein was extracted from 100 mg of 5-d-old seedlings. Seedlings were homogenized at 4°C in 500 μL modified RIPA buffer (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS) to which PMSF (250 µM), one tablet protease inhibitor cocktail (Roche), and MG132 (150 µM) were added immediately before use. Tissue lysates were centrifuged at 10,000g for 10 min at 4°C to generate a post nuclear supernatant. Then, 400 μL of the post nuclear supernatant was diluted into 600 μL wash buffer (10 mM Tris HCl, pH7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, pH 8.0, and 0.2 mM sodium orthovanadate) to which PMSF (250 µM), one tablet protease inhibitor cocktail (Roche), and MG132 (150 µM) were added immediately before use. Prior to adding antibody-conjugated beads for immunoprecipitation, an aliquot was taken for immunoblot analysis. To immunoprecipitate ubiquitinated plant proteins, 30 μL of preequilibrated antiubiquitin-conjugated agarose beads [Santa Cruz Biotechnology; catalog ub(P4D1)AC, lot J2714] was mixed with plant protein extracts and incubated for 3 h at 4°C with end-over-end rotation. Agarose beads were washed three times with 500 μL wash buffer and used for immunoblot analysis. For immunoblot analysis, proteins were run on a 7.5% polyacrylamide SDS gel and transferred to a nitrocellulose membrane (Amersham Protran; GE Healthcare Life Science). Membranes were blocked with TBS containing 0.02% Tween 20 and 5% skim milk. Membranes were incubated with primary rabbit polyclonal anti-GFP antibody (Abcam; catalog ab290, lot GR240324-1) and anti-rabbit secondary antibody (EMD Millipore; catalog AP307P, lot 2586802) at 1:2000 dilution before chemiluminescence detection using Immobilon Western Substrate (Millipore) and x-ray film (CL-X Posure; Thermo Scientific).
Protein Stability Analysis
Five-day-old seedlings were transferred to liquid one-quarter-strength MS medium containing 200 μM cycloheximide (Sigma-Aldrich) or 150 μM MG132 (Selleckchem) to inhibit new protein synthesis or to inhibit the activity of 26S proteasome, respectively. The seedlings were moved from the medium at each time point of interest and placed on filter paper to remove extra surface liquid. Ten seedlings were transferred into a 1.5-mL centrifuge tube containing 50 μL 1× Laemmli buffer and were ground with a plastic pestle. The mixture was boiled for 5 min and spun at maximum speed for 10 min to remove cell debris. The supernatant was transferred to a new tube for further immunoblot methods. Mouse anti-GFP monoclonal antibodies (1:2500; Roche; catalog 1814460, lot 10126200) were used as the primary antibody. For controls, mouse antitubulin monoclonal antibodies (1:2000; Millipore; catalog 05-661, lot 2510278) were used. Goat anti-mouse IgG-conjugated with horseradish peroxidase (1:2000; Millipore; catalog 31430, lot QD216575) was used as the secondary antibody.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CSLD2 (AT5G16910), CSLD3 (AT3G03050), CSLD5 (AT1G02730), CESA3 (AT505170), CESA6 (AT5G64740), BRI1 (AT4G39400), KNOLLE (AT1G08560), CCS52A1 (AT4G22910), CCS52A2 (AT4G11920), and CCS52B (AT5G13840).
Supplemental Data
Supplemental Figure 1. Phenotypic analysis of csld mutants
Supplemental Figure 2. Cell division is suppressed in csld5 mutants
Supplemental Figure 3. Citrine-CSLD2 and Cerulean-CSLD5 are functional fluorescent CSLD fusion proteins.
Supplemental Figure 4. Citrine-CSLD2 and eYFP-CSLD3 localize to cell plates and punctate subcellular compartments in root epidermal cells.
Supplemental Figure 5. Quantitative analysis to compare the dynamics of Cerulean-CSLD5, GFP-KNOLLE, and GFP-CESA3.
Supplemental Figure 6. Quantitative analysis of Cerulean-CSLD5 fluorescence in stomatal lineages.
Supplemental Movie 1. Dynamics of BASL in csld5 mutants.
Supplemental Movie 2. Cell plate elongation in Col-0.
Supplemental Movie 3. Cell plate elongation in csld5.
Supplemental Movie 4. Dynamics of Cerulean-CSLD5 and CYCB1;1 during cell division.
Supplemental Movie 5. Cerulean-CSLD5 colocalizes with GFP-KNOLLE during cell plate elongation.
Supplemental Movie 6. Cerulean-CSLD5 colocalizes with GFP-CESA3 during cell plate elongation.
Supplemental Movie 7. Dynamics of Cerulean-CSLD5 and GFP-KNOLLE after completion of cytokinesis.
Supplemental Movie 8. Dynamics of Cerulean-CSLD5 and GFP-CESA3 after completion of cytokinesis.
Supplemental Movie 9. Dynamics of CSLD5 after cytokinesis.
Supplementary Material
Acknowledgments
We thank Peter Doerner for sharing GFP-CYCB1;1, Gerd Juergens for sharing GFP-KNOLLE, Jianming Li for sharing BRI1-GFP and BRI1-9-GFP, and John Schiefelbein for sharing CYCB1;1-GUS transformed lines. This work was supported by grants from the Department of Energy (DE-FG02-07ER15887 to E.N.), the National Science Foundation (0937323 to E.N.), and the National Institutes of Health (RO1GM086632 to D.C.B.). M.B. was supported by a DFG postdoctoral fellowship, and D.C.B. is an investigator of the Howard Hughes Medical Institute.
AUTHOR CONTRIBUTIONS
E.N. and D.C.B. designed the research. F.G., M.B., J.R.C., and J.Y. performed the experiments. F.G., M.B., J.R.C., J.Y., and E.N. analyzed the data. F.G. and E.N. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Adrian J., et al. (2015). Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev. Cell 33: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A.J., Jensen J.K., Harholt J., Sørensen S., Moller I., Blaukopf C., Johansen B., de Lotto R., Pauly M., Scheller H.V., Willats W.G.T. (2007). Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 52: 791–802. [DOI] [PubMed] [Google Scholar]
- Bernal A.J., Yoo C.-M., Mutwil M., Jensen J.K., Hou G., Blaukopf C., Sørensen I., Blancaflor E.B., Scheller H.V., Willats W.G.T. (2008). Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 148: 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutté Y., Frescatada-Rosa M., Men S., Chow C.-M., Ebine K., Gustavsson A., Johansson L., Ueda T., Moore I., Jürgens G., Grebe M. (2010). Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J. 29: 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Durbarry A., Khatab H., Sidorova A., Doerner P., Twell D. (2009). Arabidopsis DUO POLLEN3 is a key regulator of male germline development and embryogenesis. Plant Cell 21: 1940–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R.A., Wilson S.M., Hrmova M., Harvey A.J., Shirley N.J., Medhurst A., Stone B.A., Newbigin E.J., Bacic A., Fincher G.B. (2006). Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science 311: 1940–1942. [DOI] [PubMed] [Google Scholar]
- Chen X.-Y., Kim J.-Y. (2009). Callose synthesis in higher plants. Plant Signal. Behav. 4: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-Y., Liu L., Lee E., Han X., Rim Y., Chu H., Kim S.-W., Sack F., Kim J.-Y. (2009). The Arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol. 150: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J.-C., Lerouxel O., Drakakaki G., Alonso A.P., Liepman A.H., Keegstra K., Raikhel N., Wilkerson C.G. (2007). A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA 104: 8550–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- da Silva Conceição A., Marty-Mazars D., Bassham D.C., Sanderfoot A.A., Marty F., Raikhel N.V. (1997). The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9: 571–582. [PMC free article] [PubMed] [Google Scholar]
- Davies K.A., Bergmann D.C. (2014). Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential. Proc. Natl. Acad. Sci. USA 111: 15585–15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., MacAlister C.A., Bergmann D.C. (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G. (2015). Polysaccharide deposition during cytokinesis: Challenges and future perspectives. Plant Sci. 236: 177–184. [DOI] [PubMed] [Google Scholar]
- Dwivany F.M., Yulia D., Burton R.A., Shirley N.J., Wilson S.M., Fincher G.B., Bacic A., Newbigin E., Doblin M.S. (2009). The CELLULOSE-SYNTHASE LIKE C (CSLC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Mol. Plant 2: 1025–1039. [DOI] [PubMed] [Google Scholar]
- Fagard M., Desnos T., Desprez T., Goubet F., Refregier G., Mouille G., McCann M., Rayon C., Vernhettes S., Höfte H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B., Ryan E., Foreman J., Linstead P., Boudonck K., Steer M., Shaw P., Dolan L. (2001). KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 15: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fülöp K., Tarayre S., Kelemen Z., Horváth G., Kevei Z., Nikovics K., Bakó L., Brown S., Kondorosi A., Kondorosi E. (2005). Arabidopsis anaphase-promoting complexes: multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4: 1084–1092. [PubMed] [Google Scholar]
- Galway M.E., Eng R.C., Schiefelbein J.W., Wasteneys G.O. (2011). Root hair-specific disruption of cellulose and xyloglucan in AtCSLD3 mutants, and factors affecting the post-rupture resumption of mutant root hair growth. Planta 233: 985–999. [DOI] [PubMed] [Google Scholar]
- Gavet O., Pines J. (2010). Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 18: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F., Barton C.J., Mortimer J.C., Yu X., Zhang Z., Miles G.P., Richens J., Liepman A.H., Seffen K., Dupree P. (2009). Cell wall glucomannan in Arabidopsis is synthesized by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 60: 527–538. [DOI] [PubMed] [Google Scholar]
- Guertin D.A., Trautmann S., McCollum D. (2002). Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66: 155–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman J.M., Lee J.S., Bogenschutz N.L., Peterson K.M., Virata R.E., Xie B., Kanaoka M.M., Hong Z., Torii K.U. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741. [DOI] [PubMed] [Google Scholar]
- Haga N., Kobayashi K., Suzuki T., Maeo K., Kubo M., Ohtani M., Mitsuda N., Demura T., Nakamura K., Jürgens G., Ito M. (2011). Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 157: 706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handford M.G., Baldwin T.C., Goubet F., Prime T.A., Miles J., Yu X., Dupree P. (2003). Localisation and characterisation of cell wall mannan polysaccharides in Arabidopsis thaliana. Planta 218: 27–36. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Sabatini S. (2014). Plant and animal stem cells: similar yet different. Nat. Rev. Mol. Cell Biol. 15: 301–312. [DOI] [PubMed] [Google Scholar]
- Heyman J., De Veylder L. (2012). The anaphase-promoting complex/cyclosome in control of plant development. Mol. Plant 5: 1182–1194. [DOI] [PubMed] [Google Scholar]
- Hong Z., Delauney A.J., Verma D.P.S. (2001). A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13: 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Jin H., Fitchette A.-C., Xia Y., Monk A.M., Faye L., Li J. (2009). Mutations of an alpha1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell 21: 3792–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Jin H., Tzfira T., Li J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S., Umeda M. (2011). Cell-Cycle Control and Plant Development, 1st ed. (Amsterdam, New York: Elsevier; ). [DOI] [PubMed] [Google Scholar]
- Jürgens G. (2005). Cytokinesis in higher plants. Annu. Rev. Plant Biol. 56: 281–299. [DOI] [PubMed] [Google Scholar]
- Lau O.S., Davies K.A., Chang J., Adrian J., Rowe M.H., Ballenger C.E., Bergmann D.C. (2014). Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345: 1605–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber M.H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jürgens G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139: 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A.H., Wilkerson C.G., Keegstra K. (2005). Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA 102: 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H.B., Whitney D.R. (1947). On a test of whether one of two random variables is stochastically larger than the other. In The Annals of Mathematical Statistics, Wilks S.S., ed (Baltimore, MD: Institute of Mathematical Statistics; ), pp . 50–60. [Google Scholar]
- Marrocco K., Bergdoll M., Achard P., Criqui M.-C., Genschik P. (2010). Selective proteolysis sets the tempo of the cell cycle. Curr. Opin. Plant Biol. 13: 631–639. [DOI] [PubMed] [Google Scholar]
- Menges M., Murray J.A.H. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30: 203–212. [DOI] [PubMed] [Google Scholar]
- Miart F., Desprez T., Biot E., Morin H., Belcram K., Höfte H., Gonneau M., Vernhettes S. (2014). Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant J. 77: 71–84. [DOI] [PubMed] [Google Scholar]
- Obrdlik P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101: 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M., Mastronarde D.N. (2001). Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13: 2033–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Park S., Szumlanski A.L., Gu F., Guo F., Nielsen E. (2011). A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 13: 973–980. [DOI] [PubMed] [Google Scholar]
- Richmond T.A., Somerville C.R. (2001). Integrative approaches to determining Csl function. Plant Mol. Biol. 47: 131–143. [PubMed] [Google Scholar]
- Samuels A.L., Giddings T.H. Jr., Staehelin L.A. (1995). Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J. Cell Biol. 130: 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele K., Wanner G., Kindzierski V., Jürgens G., Mayer U., Pachl F., Assaad F.F. (2009). The timely deposition of callose is essential for cytokinesis in Arabidopsis. Plant J. 58: 13–26. [DOI] [PubMed] [Google Scholar]
- Touihri S., Knöll C., Stierhof Y.-D., Müller I., Mayer U., Jürgens G. (2011). Functional anatomy of the Arabidopsis cytokinesis-specific syntaxin KNOLLE. Plant J. 68: 755–764. [DOI] [PubMed] [Google Scholar]
- Verhertbruggen Y., Yin L., Oikawa A., Scheller H.V. (2011). Mannan synthase activity in the CSLD family. Plant Signal. Behav. 6: 1620–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cnops G., Vanderhaeghen R., De Block S., Van Montagu M., Van Lijsebettens M. (2001). AtCSLD3, a cellulose synthase-like gene important for root hair growth in arabidopsis. Plant Physiol. 126: 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Schuster C., Beahan C.T., Charoensawan V., Peaucelle A., Bacic A., Doblin M.S., Wightman R., Meyerowitz E.M. (2016). Regulation of Meristem Morphogenesis by Cell Wall Synthases in Arabidopsis. Curr. Biol. 26: 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Verhertbruggen Y., Oikawa A., Manisseri C., Knierim B., Prak L., Jensen J.K., Knox J.P., Auer M., Willats W.G.T., Scheller H.V. (2011). The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol. Plant 4: 1024–1037. [DOI] [PubMed] [Google Scholar]
- Yokoyama R., Nishitani K. (2001). Endoxyloglucan transferase is localized both in the cell plate and in the secretory pathway destined for the apoplast in tobacco cells. Plant Cell Physiol. 42: 292–300. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T., Eiguchi M., Hibara K., Ito J., Nagato Y. (2013). Rice slender leaf 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J. Exp. Bot. 64: 2049–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Chen X., McCormick S. (2011). The anaphase-promoting complex is a dual integrator that regulates both MicroRNA-mediated transcriptional regulation of cyclin B1 and degradation of Cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23: 1033–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.