Figure 1.
CSLD5 Is Specifically Enriched in Dividing Stomatal Cells and Required for Stomatal Cell Division.
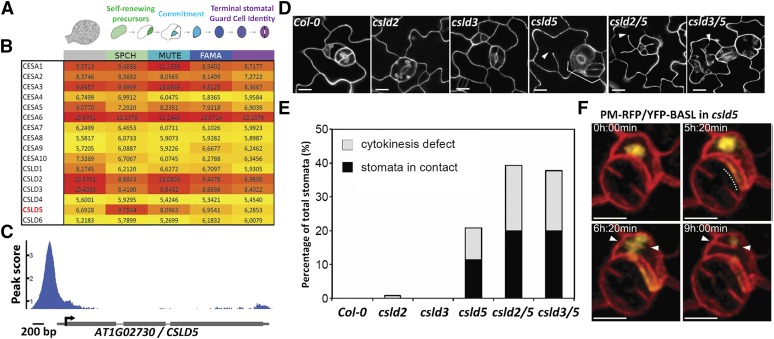
(A) Diagram of stomatal lineage cell types from which FACS-isolated populations were transcriptionally profiled (adapted from Adrian et al., 2015).
(B) Heat map of regularized and transformed (via DESeq2) log2 expression values (RNA-seq) of cell wall biosynthesis genes CesA1-10 and CSLD1-6 during stomatal lineage progression (red = highest expression, yellow = lowest; expression level ranges from 0 to 20; genes with expression values below 3 are typically not detected by reporter or microarray); CSLD5 is uniquely enriched in asymmetrically dividing meristemoids.
(C) ChIP-seq trace of SPCHpro:SPCH-Myc binding to the 5′ regulatory region of CSLD5.
(D) Confocal images of leaf epidermis 5 d after initiation cotyledons, stained with propidium iodide, showing stomatal patterning and division defects (arrowheads) in csld single and double mutants.
(E) Quantification of stomatal defects shown in (D) (n ≥ 4 leaves from ≥ 4 individuals per genotype).
(F) Time-lapse images of cells expressing a PM-RFP marker protein (mCherry-RCl2A) and the polarity protein YFP-BASL, which is correctly localized despite division defects in csld5. Dashed line indicates polarized YFP-BASL accumulation, and arrowheads indicate incomplete cell plates. Bars = 10 µm in (D) and (F).