Abstract
Programmed cell death is an integral component of Caenorhabditis elegans development. Genetic and reverse genetic studies in C. elegans have led to the identification of many genes and conserved cell death pathways that are important for the specification of which cells should live or die, the activation of the suicide program, and the dismantling and removal of dying cells. Molecular, cell biological, and biochemical studies have revealed the underlying mechanisms that control these three phases of programmed cell death. In particular, the interplay of transcriptional regulatory cascades and networks involving multiple transcriptional regulators is crucial in activating the expression of the key death-inducing gene egl-1 and, in some cases, the ced-3 gene in cells destined to die. A protein interaction cascade involving EGL-1, CED-9, CED-4, and CED-3 results in the activation of the key cell death protease CED-3, which is tightly controlled by multiple positive and negative regulators. The activation of the CED-3 caspase then initiates the cell disassembly process by cleaving and activating or inactivating crucial CED-3 substrates; leading to activation of multiple cell death execution events, including nuclear DNA fragmentation, mitochondrial elimination, phosphatidylserine externalization, inactivation of survival signals, and clearance of apoptotic cells. Further studies of programmed cell death in C. elegans will continue to advance our understanding of how programmed cell death is regulated, activated, and executed in general.
Keywords: Caenorhabditis elegans, activation phase, execution phase, programmed cell death, specification phase, WormBook
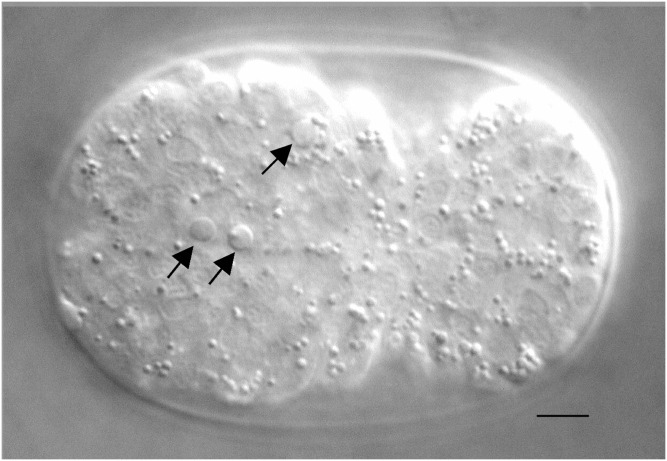
GENETIC studies of programmed cell death in Caenorhabditis elegans led to the identification of key players involved in this important physiological process, whose functions are conserved from C. elegans to humans (Adams 2003; Horvitz 2003; Danial and Korsmeyer 2004; Fuchs and Steller 2011). These pioneering studies were made possible by the following biology of C. elegans: (1) unlike in many other organisms, programmed cell death is not essential for C. elegans viability, at least under laboratory conditions (Ellis and Horvitz 1986); (2) cells undergoing programmed cell death in C. elegans change their morphology and refractivity and can be observed in living animals using differential interference contrast (DIC) microscopy (also referred to as Nomarski optics; Figure 1) (Robertson and Thomson 1982); (3) programmed cell death that occurs during the development of somatic tissues of C. elegans is determined by the essentially invariant cell lineage, therefore, it is known not only which cells undergo programmed cell death but also when and where they die (Sulston and Horvitz 1977; Sulston et al. 1983). These unique features made it possible to genetically dissect the process of programmed cell death in C. elegans at single-cell resolution. The resulting groundbreaking work was recognized with the Nobel Prize for Medicine in 2002, which was awarded to Sydney Brenner, John E. Sulston, and H. Robert Horvitz for their leading roles in deciphering the C. elegans cell lineage and in defining the genetic pathway of programmed cell death (Brenner 2003; Horvitz 2003; Sulston 2003).
Figure 1.
Nomarski image of an embryo with apoptotic cells. Three cells indicated by arrows underwent programmed cell death in a bean/comma stage embryo and exhibit a refractile, raised-button-like appearance. Bar, 5 μm.
Programmed cell death occurs during two stages of C. elegans life and in two different types of tissues: during embryonic and postembryonic development of the soma (referred to as developmental cell death) (Sulston and Horvitz 1977; Sulston et al. 1983), and in the gonad of adult hermaphrodites (germ cell death) (Sulston 1988; White 1988; Gumienny et al. 1999). Developmental cell death is determined by the essentially invariant somatic cell lineage: out of the 1090 cells generated during the development of the hermaphrodite soma, exactly 131 reproducibly undergo programmed cell death (113 of these cells die during embryonic and 18 during postembryonic development) (Sulston and Horvitz 1977; Sulston et al. 1983). Germ cell death affects the majority of all developing germ cells (possibly to provide resources for surviving germ cells) and occurs in a manner that is not determined by cell lineage (Gumienny et al. 1999; Hansen and Schedl 2013). Furthermore, various types of insults such as, for example, exposure to DNA damage-inducing treatments cause additional germ cells to die (Gartner et al. 2000). Since germ cell death has been reviewed recently (Gartner et al. 2008; Bailly and Gartner 2013), in this review we will focus on developmental cell death.
A combination of morphological observations and genetic analyses led to the finding that developmental cell death proceeds in three phases: during the “specification phase”, it is determined which cells will undergo programmed cell death and which cells will survive; during the “activation phase”, the cell death program is activated in those cells that are programmed to die; during the “execution phase”, cells are dismantled, killed, and subsequently engulfed and degraded by neighboring cells (Figure 2) (Horvitz 1999). What happens when one of these phases is disrupted? Mutations that affect the specification phase alter the highly reproducible pattern of developmental cell death and result in the inappropriate survival or death of one or a small number of cells (for example Ellis and Horvitz 1991). Mutations that affect the activation phase can cause a general block in programmed cell death (resulting in the inappropriate survival of the majority of the 131 cells that are programmed to die) or result in the inappropriate deaths of many cells that normally live (leading to the loss of viability) (Ellis and Horvitz 1986; Hengartner et al. 1992; Conradt and Horvitz 1998). Finally, mutations that disrupt the execution phase block cellular disassembly (Nakagawa et al. 2010) and result in the accumulation of dead cells (referred to as cell corpses) that fail to be engulfed and/or degraded (Sulston 1976; Hedgecock et al. 1983; Ellis et al. 1991).
Figure 2.
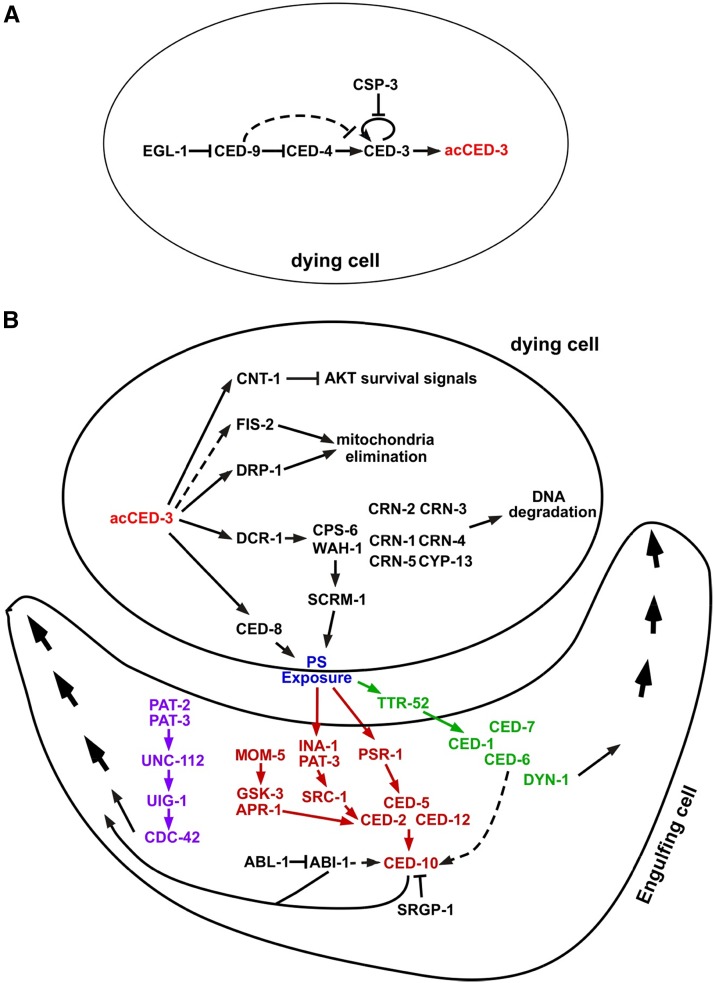
Genes involved in the activation and the execution phases of programmed cell death in C. elegans. Genes involved in two critical phases of programmed cell death, (A) activation and (B) execution, are shown. In the execution phase, four cell death execution events (fragmentation and degradation of chromosomal DNA, mitochondrial elimination, engulfment of apoptotic cell corpses, and inactivation of survival signals) are directly activated by the proteolytic cleavage of the CED-3 caspase. Three partially-redundant pathways, colored in pink, red, and green, respectively, mediate recognition and removal of apoptotic cell corpses. The activation of CED-10 and CDC-42 small GTPases leads to cytoskeletal reorganization required for pseudopod extension around an apoptotic cell. Arrows indicate confirmed activation and dashed arrows indicate proposed activation.
The first cell death abnormality (ced) genes identified were ced-1 and ced-2 (Hedgecock et al. 1983). Loss-of-function (lf) mutations in ced-1 or ced-2 partially block the engulfment of cell corpses. A block or delay in cell corpse engulfment results in the accumulation of cell corpses in embryos and young larvae, which can easily be detected using DIC microscopy. Indeed, subsequent genetic screens for mutants with similar phenotypes resulted in the identification of additional genes required for cell corpse engulfment and other aspects of the execution phase (Ellis et al. 1991; Gumienny et al. 2001; Wu et al. 2001; Zhou et al. 2001a). Mutations in ced-1 and ced-2 were also instrumental in the identification of genes involved in the activation phase. The persistent cell corpse defect in ced-1 mutants was used to screen for mutations that suppress this phenotype with the rationale that some of the suppressors should suppress this phenotype by blocking the upstream activation phase, and hence cause a general block in programmed cell death. This resulted in the identification of lf mutations in the ced-3 gene, which (as discussed in more detail below) is required for most programmed cell deaths in C. elegans (Ellis and Horvitz 1986). Genes involved in the first phase of programmed cell death, the specification phase, were identified in genetic screens with goals to identify mutations that do not cause a general block in programmed cell death, but are able to block only specific cell deaths such as, for example, the deaths of the neurosecretory motorneuron (NSM) sister cells (Ellis and Horvitz 1991; Thellmann et al. 2003; Hatzold and Conradt 2008) or the sexually dimorphic neurons cephalic companion neurons (CEMs) (Peden et al. 2007; Schwartz and Horvitz 2007). These cell death specification (ces) screens not only resulted in the identification of genes important for the specification phase, but also another crucial component of the activation phase, ced-9 (Hengartner et al. 1992). In contrast to ced-3, the function of ced-9 is to protect against programmed cell death in the 959 somatic cells that are programmed to live, and lf mutations in ced-9 cause many cells to inappropriately die; thereby leading to embryonic lethality. Does ced-3 negatively control ced-9 to allow the 131 cell deaths to occur, or does ced-9 negatively control ced-3 to allow the 959 somatic cells to survive? The activation phase of programmed cell death can be considered a regulatory pathway that controls a “life-death” switch. In such a regulatory pathway, lf mutations in genes that are closer (downstream) to the switch generally suppress the phenotype caused by lf mutations in genes that are further away (upstream) from the switch. In the case of ced-3 and ced-9, double mutant analysis revealed that the loss of ced-3 suppresses the inappropriate cell death phenotype and the embryonic lethal phenotype caused by ced-9(lf) mutations. Hence, in the regulatory pathway of the activation phase, ced-3 is epistatic to or acts downstream of ced-9 (Hengartner et al. 1992). Similar double mutant analyses have been used to place additional components of the activation phase into this life-death regulatory pathway. In addition, they have allowed analyses of genetic pathways that underlie the specification and execution phase, as described below.
Of note, rather than strictly in a sequential and linear fashion; certain aspects of the specification, activation, and execution phases may occur in parallel. Furthermore, feedback exists between the execution phase and the activation phase. This is demonstrated by the fact that in genetic backgrounds in which the activation phase is functionally compromised, a block in engulfment (execution phase) reduces the likelihood of a cell that is programmed to die to actually die (Hoeppner et al. 2001; Reddien et al. 2001; Chakraborty et al. 2015). This “killing” function of engulfment appears to act on the activation phase and promotes its swift induction and completion (Chakraborty et al. 2015).
For most of the 131 somatic cells programmed to die, the activation phase is mediated by a core apoptotic machinery that is conserved from C. elegans to mammals [egg laying defective-1 (EGL-1) homologous to the BH3-only proteins, CED-9 homologous to Bcl-2, CED-4 homologous to apoptotic protease activating factor 1 (Apaf-1), and CED-3 homologous to caspases] (Horvitz 2003; Lettre and Hengartner 2006; Conradt 2009). However, for at least one programmed cell death, the death of the “linker cell” in males, the activation phase occurs through a nonapoptotic machinery (Abraham et al. 2007; Blum et al. 2012). In the following, we will review our current understanding of the specification, activation, and execution phases of apoptotic developmental cell deaths. Furthermore, we will summarize recent advances in our understanding of the nonapoptotic death of the linker cell. Genetic perturbations and/or various treatments can also lead to various forms of nonapoptotic cell death in C. elegans (pathological cell death). Since this type of cell death does not occur during normal C. elegans development and since it has recently been reviewed (Vlachos and Tavernarakis 2010; Kinet and Shaham 2014), it will not be covered here.
Cell Death Activation
The core machinery involved in the activation of the apoptotic program
Three death-promoting genes, egl-1, ced-3, and ced-4, are required for most, if not all, developmental cell death in C. elegans. Strong lf mutations in any of these genes result in the survival of most somatic cells that normally undergo programmed cell death during development (Ellis and Horvitz 1986; Conradt and Horvitz 1998). Furthermore, these three genes act within dying cells to promote apoptosis, indicating that cells die by an intrinsic suicide mechanism (Yuan and Horvitz 1990; Shaham and Horvitz 1996b; Conradt and Horvitz 1998). By contrast, the activity of the ced-9 gene protects cells from undergoing programmed cell death during C. elegans development (Hengartner et al. 1992). lf mutations in ced-9 cause embryonic lethality as a consequence of ectopic deaths of cells that normally live. ced-3, ced-4, egl-1, and ced-9 appear to act in a simple genetic pathway in which egl-1 acts upstream of ced-9 to induce cell death, ced-9 acts upstream of ced-4 to inhibit cell death, and ced-4 acts upstream of ced-3 to kill cells (Figure 2A) (Hengartner et al. 1992; Shaham and Horvitz 1996b; Conradt and Horvitz 1998).
ced-9 encodes a protein similar to the gene product of the human proto-oncogene bcl-2 (Hengartner and Horvitz 1994b), which plays a similar role in preventing apoptosis in mammals (Adams and Cory 2001). ced-9 and bcl-2 are members of a gene family that plays important roles in regulating apoptosis in diverse organisms (Reed 1997; Adams and Cory 2001). Members of the Bcl-2 protein family contain one or several characteristic Bcl-2 homology (BH) domains, BH1, BH2, BH3, and BH4, which are domains important for mediating interactions among different members of the Bcl-2 family (Adams and Cory 2001). egl-1 encodes a small protein of 91 amino acids with a BH3 motif, which has been found in all proapoptotic members of the Bcl-2 gene family and mediates direct binding of these proteins to antipoptotic Bcl-2 members (Conradt and Horvitz 1998; Bouillet and Strasser 2002). ced-3 encodes the founding member of a family of aspartate-specific cysteine proteases named caspases (Yuan et al. 1993; Alnemri et al. 1996). Like other caspases, CED-3 is synthesized as a proenzyme and is proteolytically activated to generate an active protease containing a large subunit of 17 kDa (p17) and a small subunit of 15 kDa (p15) or 13 kDa (p13) (Alnemri et al. 1996; Xue et al. 1996). The protease activity of CED-3 appears to be essential for ced-3 to cause programmed cell death in C. elegans (Xue et al. 1996; Shaham et al. 1999). However, a deletion mutation that removes the ced-3 region encoding the entire protease domain, including the p17 and p15 domains, causes a weaker cell death defect than those observed in multiple ced-3(lf) mutants carrying missense mutations (Shaham et al. 1999), suggesting that some of the developmental cell death can occur in the absence of the CED-3 protease activity. ced-4 encodes a protein similar to human Apaf-1, an activator of human caspase-9 (Yuan and Horvitz 1992; Zou et al. 1997). Both CED-4 and Apaf-1 contain a caspase-recruitment domain and nucleotide-binding motifs that are critical for the function of these proteins (Seshagiri and Miller 1997; Zou et al. 1999). Likewise, CED-4 plays a critical role in activating CED-3 during apoptosis. Interestingly, ced-4 may also produce an alternatively spliced transcript, ced-4L, which encodes a slightly larger protein (CED-4L) with a 24-amino acid insertion between its two nucleotide-binding motifs and which might protect against programmed cell death (Shaham and Horvitz 1996a). The serine/arginine-rich (SR) protein kinase 1 (spk-1) gene, which encodes a homolog of SR protein kinases implicated in regulating splicing, has been proposed to inhibit cell death in C. elegans by promoting the generation of the ced-4L splice variant (Galvin et al. 2011). Consistently, loss of spk-1 preferentially suppresses the cell death defects of some partial ced-4(lf) mutants but not those of strong ced-4(lf) mutants.
Biochemical and structural analyses of the activation of the core apoptotic program
Biochemical, cell biological, and structural analyses of EGL-1, CED-9, CED-4, and CED-3 have provided important insights into how these proteins function to regulate the activation of programmed cell death during C. elegans development (Horvitz 2003). CED-4 has been shown to physically interact with CED-9 in vitro and in cultured cells (Chinnaiyan et al. 1997; Spector et al. 1997; Wu et al. 1997), forming a 2:1 CED-4/CED-9 protein complex (Yan et al. 2005). In vivo, endogenous CED-9 and CED-4 proteins have been shown to colocalize at mitochondria in C. elegans embryos, and the mitochondrial localization of CED-4 appears to be dependent on CED-9 (Chen et al. 2000). In addition to CED-9, CED-4 has been shown to interact with CED-3 in vitro and in mammalian cells (Chinnaiyan et al. 1997; Yang et al. 1998).
Interestingly, ectopic egl-1 expression in C. elegans embryos results in the translocation of CED-4 to perinuclear membranes and ectopic programmed cell death (Chen et al. 2000). CED-4 translocation from mitochondria to perinuclear membranes appears to be triggered by the binding of EGL-1 to CED-9, which induces a major conformational change in the CED-9 protein (Yan et al. 2004), resulting in the disassociation of the CED-4 dimer from the CED-4/CED-9 complex (Conradt and Horvitz 1998; del Peso et al. 1998; Parrish et al. 2000; Yan et al. 2005). The released CED-4 dimers then oligomerize to form a funnel-shaped CED-4 octamer, which may recruit two CED-3 zymogens and facilitate its autocatalytic activation through zymogen dimerization (Qi et al. 2010; W. Huang et al. 2013). Moreover, this series of events can be recapitulated in vitro using recombinant EGL-1, CED-4, and CED-9 protein; leading to the proteolytic activation of the CED-3 zymogen (Yan et al. 2005). Therefore, these four proteins are necessary and sufficient to activate CED-3 in vitro.
A gain-of-function mutation in ced-9 (n1950) results in the substitution of glycine 169 with glutamate, blocks most somatic cell death during development (Hengartner and Horvitz 1994a), and impairs the binding of EGL-1 to CED-9 and EGL-1-induced release of the CED-4 dimers (Parrish et al. 2000; Chen et al. 2000; Yan et al. 2004, 2005). EGL-1-induced CED-4 disassociation from CED-9 and its translocation to perinuclear membranes are thought to be important for the activation of CED-4 and the subsequent activation of the CED-3 zymogen (Chen et al. 2000; Yan et al. 2005). CED-4 translocation to perinuclear membranes may help stabilize CED-4 octamers or help facilitate the interaction between CED-4 octamers and the CED-3 zymogens. However, the subcellular localization pattern of the CED-3 zymogen and the mechanism that relocates CED-4 to perinuclear membranes have not been determined and are critical for understanding cell death activation in C. elegans. A recent study proposes that CED-4 predominantly localizes to perinuclear membranes in living cells and further accumulates on perinuclear membranes in response to apoptotic stimuli in a manner dependent on EGL-1 (Pourkarimi et al. 2012). It is unclear why different CED-4 antibodies used in these two studies exhibited drastically different CED-4 localization patterns (Chen et al. 2000).
The mechanism by which CED-3 is activated appears to differ somewhat from the mechanisms that activate mammalian caspases, which involve either release of cytochrome c from mitochondria and assembly of an oligomerized Apaf-1/caspase-9 apoptosome (caspase-9 activation), the formation of caspase-8 trimers induced by activation of death receptors (caspases-8 activation), or direct proteolytic activation of downstream executor caspases (such as caspase-3, caspase-6, and caspase 7) by upstream initiator caspases (such as caspase-8 and caspase-9) (Liu et al. 1996; Budihardjo et al. 1999; Jiang and Wang 2004).
Although CED-9 clearly serves as a cell death inhibitor, some genetic evidence suggests that ced-9 also has a proapoptotic activity (Hengartner and Horvitz 1994a). In a partial ced-3(lf) mutant background, loss of ced-9 can significantly enhance the cell death defect of the ced-3 mutant. It is unclear if ced-9 generates a different transcript that encodes a proapoptotic protein. Alternatively, the proapoptotic activity of CED-9 could be due to its ability to act as a chaperone to assemble an asymmetric CED-4 dimmer, which is required for the formation of the proapoptotic CED-4 octamers (Yan et al. 2005; Qi et al. 2010). It has also been suggested that CED-9 may promote cell killing by promoting mitochondrial fragmentation (Jagasia et al. 2005).
Regulation of cell death activation in C. elegans
Given the cell killing function of CED-3, it is critical that the killing activity of CED-3 be tightly regulated. The control of caspase activity can be achieved at two different levels: the activation of the caspase zymogens and the catalytic activity of activated caspases. In mammals, inhibitors of apoptosis (IAPs) directly suppress both the activation of caspase zymogens and the catalytic activity of activated caspases (Budihardjo et al. 1999; Riedl and Shi 2004). Intriguingly, no IAP homolog has been identified in C. elegans, suggesting that different caspase inhibitors are employed to negatively regulate the activation or the activity of the CED-3 caspase.
There are three genes in C. elegans encoding caspase-like proteins: caspase (csp) 1, csp-2, and csp-3 (Shaham 1998). Two of the caspase-like proteins, CSP-2 and CSP-3, appear to lack a caspase activity in vitro. CSP-3 is a smaller protein that shares sequence similarity with the small subunit of the CED-3 caspase and is not expected to act as a functional caspase. Although CSP-2 has an overall sequence similarity to the protease domain of the CED-3 caspase, it lacks the invariant catalytic pentapeptide QACXG (C is the active site and X could be R, Q, or G) that is found in all active caspases (VCCRG in CSP-2) (Cohen 1997; Geng et al. 2008; Geng et al. 2009). CSP-2 and CSP-3 may thus act dominant-negatively to interfere with the activation or the activity of CED-3. Indeed, both CSP-2 and CSP-3 can associate with the CED-3 zymogen and inhibit CED-3 autocatalytic activation in vitro (Geng et al. 2008, 2009). However, CED-4 oligomers can overcome the inhibitory effects of CSP-2 and CSP-3 to activate the CED-3 zymogen, providing a mechanism by which CED-3 is only activated in dying cells where CED-4 is activated and is inhibited in cells that are not programmed to die. Consistent with these in vitro observations, inactivation of the csp-2 and csp-3 gene in C. elegans causes ectopic cell death in germ cells and somatic cells, respectively (Geng et al. 2008, 2009; Huang et al. 2012). Therefore, CSP-2 and CSP-3 employ the same mechanism to prevent undesired caspase zymogen autoactivation and apoptosis in different tissues of C. elegans and define a new class of caspase inhibitors that act at the level of preventing caspase zymogen autoactivation. The cell death inhibitory effects of CSP-2 and CSP-3 appear to be quite weak, since the effect of csp-3 on cell death was not observed in another study (Denning et al. 2013). There are probably additional caspase inhibitors acting in parallel. One potential caspase inhibitor is CED-9, which is an excellent CED-3 substrate (Xue and Horvitz 1997). CED-9 has been shown to act as a competitive inhibitor of CED-3 (Xue and Horvitz 1997), as alterations of two CED-3 cleavage sites in CED-9 markedly impair its cell death inhibitory activity. Unlike CSP-2 and CSP-3, the third caspase homolog, CSP-1, does show caspase activity in vitro, which has a different substrate specificity from that of CED-3 (Shaham 1998), and has been shown to have a weak proapoptotic activity in some specific cells; acting independently of the core apoptotic pathway, including CED-4 and CED-9 (Denning et al. 2013).
In addition to egl-1, ced-3, ced-4, and ced-9, several other genes have been implicated in regulating the activation of the apoptotic program during C. elegans development. These include the defender against apoptotic death 1 (dad-1) gene (Sugimoto et al. 1995), which encodes a protein similar to the mammalian apoptosis inhibitor DAD1 (Nakashima et al. 1993); the inhibitor of cell death 1 (icd-1) gene, which encodes a protein similar to the β subunit of the nascent polypeptide-associated complex (Bloss et al. 2003); the dynamin-related protein 1 (drp-1) gene, which encodes a dynamin GTPase related protein that mediates mitochondrial fission (Jagasia et al. 2005); the adenine nucleotide translocator 1.1 (ant-1.1) gene (also called wan-1), which encodes a homolog of the human adenine nucleotide translocator (Shen et al. 2009); and the eukaryotic initiation factor 3 subunit K (eif-3.K) gene, which encodes a homolog of eif-3.k (Huang et al. 2012). Both DRP-1 and ANT-1.1 localize to mitochondria and are thought to interact with CED-9 and/or CED-4 (for ANT-1.1) to affect apoptosis (Jagasia et al. 2005; Shen et al. 2009; Y. Lu et al. 2011). eif-3.K appears to act upstream of ced-3 to promote apoptosis (Huang et al. 2012). How dad-1 and icd-1 might interact with the core killing machinery is currently unclear.
Cell Death Specification
The observation that cell fate-altering mutations, such as lf mutations of the genes uncoordinated 86 (unc-86) POU or pattern of reporter gene expression abnormal 3 (pag-3) Gfi-1, can affect the pattern of developmental cell death suggests that programmed cell death which occurs during the development of the C. elegans soma can be regarded as a cell fate (Chalfie et al. 1981; Sulston and Horvitz 1981; Finney et al. 1988; Cameron et al. 2002). Furthermore, most of the 131 cells that die are generated through a cell division that is asymmetric with respect to both cell fate and cell size (with the smaller daughter being the cell that is programmed to die) and die in a cell-autonomous manner (Sulston and White 1980; Yuan and Horvitz 1990). This suggests that these cells “know” at the time of their birth that their fate is to die and, hence, are indeed programmed to die. Finally, many of the 131 cells that die are sisters of cells that differentiate into neurons and adopt a neuronal fate, if prevented from dying (Ellis and Horvitz 1986; Ellis and Horvitz 1991; White et al. 1991). At least some of these “undead” neurons appear to be fully functional (Avery and Horvitz 1987).
egl-1 is the key activator of the activation phase of apoptotic cell death. The current model for what determines which cells will live and which cells will die during development is that in the 959 cells programmed to live, the activity of egl-1 is low or absent and that in the 131 cells that are programmed to die, egl-1 activity is high. High egl-1 activity inhibits ced-9 activity, resulting in the activation of ced-4 and ced-3 and the induction of the execution phase of apoptotic cell death (Horvitz 2003). Therefore, during the specification phase of apoptotic cell death, the activity of egl-1 has to be increased specifically in those cells that are programmed to die.
Role of egl-1 transcriptional control
egl-1 activity is regulated at the level of transcription. The egl-1 gene is expressed at a detectable level predominantly in cells programmed to die (Conradt and Horvitz 1999; Thellmann et al. 2003; Liu et al. 2006; Hatzold and Conradt 2008; Potts et al. 2009; Hirose et al. 2010; Winn et al. 2011; Hirose and Horvitz 2013; Jiang and Wu 2014; Wang et al. 2015). Furthermore, mutations in cis-regulatory elements of the egl-1 locus not only cause changes in egl-1 expression but also in the pattern of programmed cell death (Conradt and Horvitz 1999; Hirose et al. 2010). These cis-regulatory elements are located either downstream or upstream of the egl-1 transcription unit and are conserved in related Caenorhabditis species (Figure 3A). A number of direct transcriptional regulators (trans-acting factors) of the egl-1 gene that act through these elements have been identified (Table 1) (Conradt and Horvitz 1999; Thellmann et al. 2003; Liu et al. 2006; Hatzold and Conradt 2008; Potts et al. 2009; Hirose et al. 2010; Winn et al. 2011; Hirose and Horvitz 2013; Jiang and Wu 2014; Wang et al. 2015). Their genetic analyses revealed that most control egl-1 transcription (and, hence, programmed cell death) only in one type of cell lineage, or a limited number of different types of cell lineages. Furthermore, most of these transcriptional regulators act through one specific cis-regulatory element of the egl-1 locus (Figure 3A). Hence, egl-1 transcriptional control appears to be mediated by a composite of lineage-specific modules. In support of this notion, analyses of known regulators of egl-1 transcription suggest that many of them have additional, nonapoptotic functions, including nonapoptotic functions in the particular lineage or lineages in which they control egl-1 transcription. Nevertheless, the functions of these transcriptional regulators in the regulation of apoptotic cell death appear to be conserved since most of them have human homologs that have been implicated in the regulation of apoptotic cell death and/or tumorigenesis (Potts and Cameron 2011).
Figure 3.
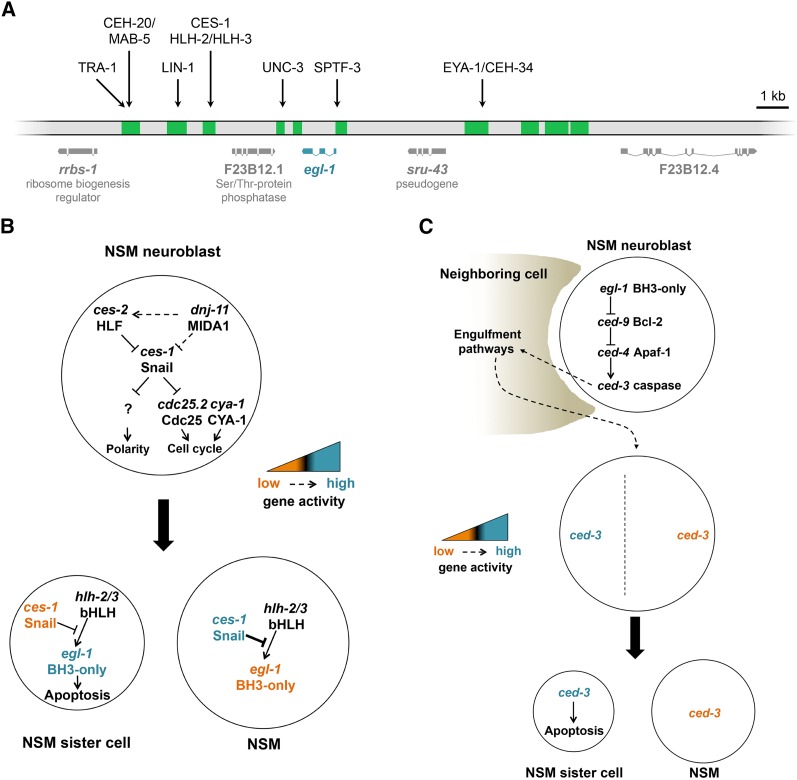
Cell death specification. (A) cis-elements and transcription factors regulating the transcription of egl-1. egl-1 transcription unit as well as transcription units located upstream and downstream of the egl-1 transcription unit are shown. The gray bar represents part of chromosome V and green regions represent sequences that are outside of coding regions and that are conserved in other Caenorhabditis species. Many of these regions have been shown to be required for egl-1 transcriptional control in specific cells or lineages, and hence, represent cis-regulatory elements of the egl-1 locus. Indicated above the schematic of the egl-1 locus are transcription factors that have been shown to directly control egl-1 transcription by binding to specific cis-regulatory elements. (B) Model for the transcriptional upregulation of egl-1 in the NSM sister cell. In the NSM neuroblast, the activity of the Snail-like gene ces-1 is negatively controlled by the HLF-like transcription factor gene ces-2 and the MIDA1-like gene dnj-11. ces-1 activity in the NSM neuroblast can affect NSM neuroblast polarity through targets of ces-1 that are currently unknown. It also can affect cell cycle progression by suppressing the activity of the cdc-25.2 gene, which is required for cell cycle progression. After the asymmetric division of the NSM neuroblast, ces-1 activity is detected in the larger NSM but not in the smaller NSM sister cell. This asymmetry in ces-1 results in the ces-1-dependent repression of egl-1 transcription in the NSM but not in the NSM sister cell. In the NSM sister cell, egl-1 transcription can occur and this is dependent on the HLH genes hlh-2 and hlh-3. (C) Model for the killing function of the engulfment pathways in the NSM lineage. In the NSM neuroblast, the central cell death pathway is activated to a certain degree through a mechanism that remains to be determined. ced-3 activity generated in the NSM neuroblast leads to the activation of the engulfment pathways in neighbors of the NSM neuroblast. The engulfment pathways subsequently promote the polarization of the NSM neuroblast and the formation of a gradient of ced-3 activity along the cell division axis. As a result of this gradient, the smaller NSM sister cell inherits more ced-3 activity than the larger NSM and this facilitates the killing of the NSM sister cell.
Table 1. Direct regulators of egl-1 BH3-only transcription.
| Transcriptional regulators | Cells | Function in egl-1 transcription | Reference |
|---|---|---|---|
| TRA-1 | HSN | repressor | Conradt and Horvitz 1999 |
| HLH-2, HLH-3 | NSM sister | activator | Thellmann et al. 2003 |
| CES-1 | NSMa | repressor | Thellmann et al. 2003 |
| CEH-20, MAB-5 | P11.aaap | activator | Liu et al. 2006 |
| CEH-20, LIN-39, UNC-62 | VC neurons | repressor | Potts et al. 2009 |
| CEH-34, EYA-1 | M4 sister cell | activator | Hirose et al. 2010 |
| EFL-3, LIN-39 | VA and VB neurons | repressor | Winn et al. 2011 |
| SPTF-3 | M4 sister cella | activator | Hirose and Horvitz 2013 |
| LIN-1 | g1A sister, I1 sistera | activator | Jiang and Wu 2014 |
| UNC-3 | RID sister | activator | Wang et al. 2015 |
Direct regulators of egl-1 transcription identified that either function as repressors or activators of egl-1 transcription in specific cells.
Indicates that this particular transcriptional regulator affects the programmed death of additional cells.
RID lineage:
The Collier/Olf1/EBF1 (COE) transcription factor UNC-3 plays such a dual role in the RID lineage (Wang et al. 2015). The RID progenitor gives rise to the RID, which differentiates into a neuron; and the RID sister cell, which dies (Sulston et al. 1983). unc-3 expression is detected in all three cells, the RID progenitor, the RID neuron, and the RID sister cell. In the RID neuron, the loss of unc-3 function results in a defect in certain aspects of neurite growth; in the RID sister cell, however, the loss of unc-3 prevents its programmed death (Wang et al. 2015). The mechanism through which the UNC-3 protein affects neurite growth in the RID neurons is still unknown. However, in the RID sister cell, the UNC-3 protein activates programmed cell death by binding to a cis-regulatory region of the egl-1 locus located downstream of the egl-1 transcription unit, thereby directly activating egl-1 transcription in the RID sister cell (Figure 3A) (Wang et al. 2015). What remains to be determined is through what mechanism UNC-3-dependent activation of egl-1 transcription is prevented in the RID neuron, which is programmed to survive.
The deaths of the hermaphrodite-specific neurons and cephalic companion neurons:
Two types of programmed cell deaths occur in a sexually dimorphic manner during C. elegans development: the deaths of the two hermaphrodite-specific neurons (HSNs) in males and the deaths of the four CEMs in hermaphrodites (Sulston et al. 1983). The Zn finger DNA-binding protein and transcriptional repressor transformer 1 (TRA-1) Gli, which was originally identified because of its role in sex determination (Hodgkin 1987; Zarkower and Hodgkin 1992), plays a critical role in the specification of the HSN and CEM death. TRA-1 functions as the terminal, global regulator of somatic sexual fate and, hence, specifies the development of sexually dimorphic features, including the sexually dimorphic presence of the HSNs and CEMs. In hermaphrodites, in which TRA-1 activity is high, TRA-1 binds to another downstream cis-regulatory element of the egl-1 locus; thereby directly repressing egl-1 transcription in the HSNs to allow HSN survival in hermaphrodites (Figure 3A) (Conradt and Horvitz 1999). However, by directly repressing the transcription of the gene C. elegans homeobox 30 (ceh-30), which encodes a BarH homeodomain transcription factor that may directly repress egl-1 transcription in the CEMs, TRA-1 indirectly activates egl-1 transcription in the CEMs to cause CEM death in hermaphrodites (Peden et al. 2007; Schwartz and Horvitz 2007). In males, in which TRA-1 activity is low, TRA-1 is unable to repress egl-1 transcription in the HSNs and ceh-30 transcription in the CEMs; consequently, the HSNs die and the CEMs survive.
Role of asymmetric cell division
As mentioned above, most of the 131 cells that die during development are generated through a cell division that is asymmetric with respect to both cell fate and cell size (Sulston and Horvitz 1977; Sulston et al. 1983). Furthermore, in those lineages in which egl-1 transcription has been analyzed, egl-1 transcription can specifically be detected in the daughter that is programmed to die (Conradt and Horvitz 1999; Thellmann et al. 2003; Liu et al. 2006; Hatzold and Conradt 2008; Potts et al. 2009; Hirose et al. 2010; Winn et al. 2011; Hirose and Horvitz 2013; Jiang and Wu 2014; Wang et al. 2015). How is this asymmetry in egl-1 transcriptional activation and, hence, egl-1 activity achieved? It has been proposed that this is achieved through the asymmetric presence or activity in the two daughter cells of activators and/or repressors of the cell death fate and egl-1 transcription (Guenther and Garriga 1996; Frank et al. 2005; Hatzold and Conradt 2008; Chien et al. 2013). Indeed, mutations that affect the abilities of mothers of cells programmed to die to divide asymmetrically can affect the cell death fate of their daughters (Guenther and Garriga 1996; Cordes et al. 2006; Hatzold and Conradt 2008; Ou et al. 2010; Singhvi et al. 2011; Chien et al. 2013; Gurling et al. 2014; Teuliere et al. 2014). Therefore, events that lead to the polarization of mothers of cells programmed to die and that are required for their abilities to divide asymmetrically are fundamentally important for cell death specification. These events most probably include cell nonautonomous signaling events. One signaling pathway that has recently been implicated in the regulation of cell death is the C. elegans LET-60 Ras and MAP kinase 1 (MPK-1) ERK MAPK pathway. It was shown that the EGF-like ligand abnormal cell lineage 3 (LIN-3) can promote a number of developmental cell deaths, such as the deaths of the g1A sister cell and the I1 sister cell (Jiang and Wu 2014). In this context, LIN-3 activates the LET-60 Ras and MPK-1 ERK MAPK pathway, which results in the binding of the ETS-like transcription factor LIN-1 to a downstream cis-regulatory region of the egl-1 locus and LIN-1-dependent activation of egl-1 transcription (Figure 3A) (Jiang and Wu 2014). Based on these results it has been proposed that LIN-3-dependent signaling contributes to the activation of the core apoptotic machinery in a cell-nonautonomous manner by promoting egl-1 transcriptional upregulation in cells programmed to die. An alternative explanation could be that LIN-3-dependent signaling may promote the polarization of mothers of cells programmed to die (and their abilities to divide asymmetrically) and prime the egl-1 locus for transcriptional upregulation in the smaller daughter after cell division.
The Q lineage:
A number of genes have been identified that are required for the asymmetric divisions of the left and right posterior daughter of the Q cell (Q.p) during the first larval stage (L1 stage) and the apoptotic deaths of their smaller daughters (Cordes et al. 2006; Ou et al. 2010; Singhvi et al. 2011; Chien et al. 2013; Gurling et al. 2014; Teuliere et al. 2014). [Q.pL and Q.pR divide asymmetrically and each gives rise to a larger daughter, Q.pa, which further divides to generate two neurons (AVM/PVM and SDQL/SDQR, respectively), and a smaller daughter, Q.pp, which dies (Sulston and Horvitz 1977).] These genes define three pathways that contribute to the ability of Q.p to divide asymmetrically by size and/or fate: the Par-1-like gene 1 (pig-1) pathway (Cordes et al. 2006; Chien et al. 2013), the ADP-ribosylation factor (arf) pathway (Singhvi et al. 2011; Teuliere et al. 2014), and a target of ERK kinase MPK-1 (toe-2)-dependent pathway (Gurling et al. 2014). The pig-1 pathway is comprised of the genes abnormal embryonic partitioning of cytoplasma 4 (par-4) LKB1, pig-1 MELK, yeast STE20 related adaptor protein homolog 1 (strd-1) STRAD, and the mouse embryo scaffolding protein homolog mop-25.2 MOP25. The arf pathway includes the genes arf-1.2 Arf, arf-6 Arf, centaurin 2 (cnt-2) Arf GTPase-activating protein (GAP), general receptor for phosphoinositides 1 (grp-1) Arf guanine nucleotide exchange factor (GEF), exchange factor for Arf 6 (efa-6) Arf GEF, BRag/Iqsec/Schizo related Arf GEF family member 1 (bris-1) Arf GEF, RAB family 5 (rab-5) Rab5, and dynamin related 1 (dyn-1) Dynamin. Many of these genes affect additional asymmetric cell divisions that give rise to cells that are programmed to die, such as the division of Q.a, and divisions in the PLM/ALN and HSN/PHB lineages. The pig-1 pathway potentially also affects the asymmetric divisions of cells that give rise to some of the first 14 cell deaths that occur during development as well as the M4 mother cell that gives rise to the M4 sister cell, which is programmed to die (see below: The M4 sister cell death) (Denning et al. 2012; Hirose and Horvitz 2013). The identification of these pathways suggests that events at the plasma membrane as well as membrane trafficking events (potentially in an endocytic compartment) play an important role in the polarization of Q.p and its ability to divide asymmetrically. What remains to be determined is how these three pathways interact, what signal or signals they help to transduce, and how their functions relate to the programming of the cell death fate and the activation of egl-1 transcription in the smaller daughter cell, in particular (Guenther and Garriga 1996; Frank et al. 2005; Cordes et al. 2006; Hatzold and Conradt 2008; Ou et al. 2010; Singhvi et al. 2011; Chien et al. 2013; Gurling et al. 2014; Teuliere et al. 2014).
The NSM sister cell death:
In the NSM lineage, transcription factors have been identified that functionally connect the asymmetric division of a mother cell with asymmetric egl-1 transcription in the daughters (Figure 3B). The NSM neuroblast divides asymmetrically to give rise to a larger daughter, the NSM, which differentiates into a serotonergic neuron; and a smaller daughter, the NSM sister cell, which dies (Sulston et al. 1983; Hatzold and Conradt 2008). It has been proposed that egl-1 transcription can potentially be activated in the NSM and the NSM sister cell through a heterodimer of helix-loop-helix 2 (HLH-2) Daughterless and HLH-3 Achaete scute (HLH-2/HLH-3), which is present in both daughter cells and which can bind to four E boxes/Snail binding sites located in a downstream cis-regulatory element of the egl-1 locus (Figure 3A) (Thellmann et al. 2003). However, HLH-2/HLH-3-dependent activation of egl-1 transcription appears to be prevented in the larger daughter, the NSM, by the Snail-like Zn-finger transcription factor CES-1, which can only be detected in the NSM but not the NSM sister cell (Ellis and Horvitz 1991; Metzstein and Horvitz 1999; Thellmann et al. 2003; Hatzold and Conradt 2008). CES-1 prevents egl-1 transcription by competing with HLH-2/HLH-3 for binding to the same E boxes/Snail binding sites. The mechanism or mechanisms through which the asymmetric presence of CES-1 protein in the daughter cells is achieved remains to be determined; however, ces-1 function itself appears to contribute to the ability of the NSM neuroblast to divide asymmetrically. Mutations that result in the mis- or overexpression of the ces-1 gene in the NSM neuroblast cause the NSM neuroblast to divide symmetrically giving rise to two daughter cells of similar sizes, both containing detectable levels of CES-1 (Hatzold and Conradt 2008). Mutations that cause ces-1 mis- or overexpression in the NSM neuroblast are lf mutations of the genes ces-2 and DNaJ domain 11 (dnj-11), which encode a hepatic leukemia factor (HLF)-like bZIP transcription factor and a Mida1/ZRF1-like chaperone, respectively; and a gain-of-function mutation of ces-1, which is located in a cis-regulatory region of the ces-1 locus (Ellis and Horvitz 1991; Metzstein et al. 1996; Metzstein and Horvitz 1999; Hatzold and Conradt 2008). Consequently, egl-1 transcription is repressed in both daughters and both daughters survive. Mis- or overexpression of the ces-1 gene in the NSM neuroblast also compromises cell cycle progression in this cell, which suggests that the correct level of CES-1 in the NSM neuroblast is not only critical for the ability of this cell to divide asymmetrically but also for its ability to divide at the correct time (Yan et al. 2013). It has been proposed that CES-1 exerts its effect on cell cycle progression in the NSM lineage by directly controlling the transcription of the gene cell division cycle 25.2 (cdc25.2), which encodes a CDC25-like phosphatase that promotes cell cycle progression (Yan et al. 2013). How CES-1 affects the polarization of the NSM neuroblast is still unclear (Figure 3B). Interestingly, the cell death regulatory function of the CES-2 HLF, CES-1 Snail, and EGL-1 BH3-only pathway appears to be conserved in mammals. HLF and the Snail-related Zn-finger transcription factor SLUG act in the hematopoietic lineage in mammals to regulate the transcription of the BH3-only gene Puma (Inaba et al. 1996; Inukai et al. 1999; Inoue et al. 2002; Wu et al. 2005). Finally, as mentioned in the Introduction, the pathways that mediate cell corpse engulfment also have a killing function (Hoeppner et al. 2001; Reddien et al. 2001). This killing function was recently investigated in the NSM lineage. Based on this study it has been proposed that the engulfment pathways promote the death of the NSM sister cell by contributing to the polarization of the NSM neuroblast; the generation of a gradient of “apoptotic potential” (including active CED-3 caspase and the potential to synthesize additional CED-3 protein) in the NSM neuroblast; and the asymmetric segregation of apoptotic potential into the smaller daughter, which is programmed to die (Figure 3C) (Chakraborty et al. 2015). The study also revealed that the activation of the engulfment pathways in this context is induced through the core apoptotic machinery, which appears to already be active to a certain degree in the NSM neuroblast (Figure 3C). However, many questions remain, including the mechanism through which the core apoptotic machinery is tightly controlled in the NSM neuroblast and the nature of the signaling pathways involved. Furthermore, it is currently unclear whether the engulfment pathways play a similar role in other cell lineages.
The M4 sister cell death:
The M4 mother divides to give rise to M4, which differentiates into a motor neuron; and the M4 sister cell, which dies (Sulston et al. 1983). The death of the M4 sister cell is controlled by a heterodimer composed of the homeodomain-containing transcription factor CEH-34 and the Eyes absent-like (EYA) transcription factor EYA-1 (EYA homolog), CEH-34/EYA-1. CEH-34/EYA-1 binds to an upstream cis-regulatory element of the egl-1 locus (Figure 3A), thereby directly activating the transcription of egl-1 in the M4 sister cell (Hirose et al. 2010). The death of the M4 sister cell and the upregulation of egl-1 transcription in the M4 sister cell are also at least partially dependent on the specificity protein 1 (SP1)-like transcription factor SPTF-3, which binds to another upstream cis-regulatory element of the egl-1 locus (Figure 3A) (Hirose and Horvitz 2013). Interestingly, SPTF-3 is a direct transcriptional activator of both the egl-1 gene and the pig-1 gene, which encodes an AMPK-like protein kinase most similar to the mammalian kinase MELK and which, as discussed above (see The Q lineage), is a component of a genetic pathway that has been implicated in asymmetric cell division (by size and fate) in a number of cell lineages. Consistent with the notion that SPTF-3 activates pig-1 transcription in the M4 lineage to cause the death of the M4 sister cell; like the loss of sptf-3, the loss of pig-1 blocks the death of ∼50% of the M4 sister cells (Hirose and Horvitz 2013). Based on these findings it was proposed that the death of the M4 sister cell is controlled by two parallel pathways that are both induced by sptf-3 function: the core apoptotic cell death pathway that is activated by the sptf-3-, ceh-34-, and eya-1-dependent transcriptional upregulation of egl-1 (the sptf-3, ceh-34, eya-1, egl-1 pathway) and a pathway that is independent of the core apoptotic pathway and that is activated by sptf-3-dependent transcriptional activation of pig-1 (the sptf-3, pig-1 pathway). Finally, the death of the M4 sister cell (as well as a number of other programmed cell deaths, including the death of the NSM sister cells) is also at least partially dependent on the yeast general control nondepressible homolog gcn-1 gene and the ABC transporter, class F 1 (abcf-1) gene, whose gene products physically interact and, based on sequence homologies, may function in the regulation of messenger RNA translation (Hirose and Horvitz 2014). This pathway (gcn-1, abcf-1 pathway) has been proposed to act in parallel to the two sptf-3-dependent pathways to contribute to the death of the M4 sister cell as well (Hirose and Horvitz 2014). However, based on the known function of pig-1 in asymmetric cell division and recent findings in the NSM lineage (Cordes et al. 2006; Chien et al. 2013; Chakraborty et al. 2015), it is also possible that, rather than acting in parallel to the sptf-3, ceh-34, eya-1, egl-1 pathway; the sptf-3, pig-1 pathway and the gcn-1, abcf-1 pathway may act at different time points in the M4 lineage to promote the asymmetric division of the M4 mother cell and the segregation of the apoptotic potential into the M4 sister cell (sptf-3, pig-1 pathway) as well as the synthesis of CED-3 protein in the M4 sister cell after cell division (gcn-1, abcf-1 pathway).
Noncanonical apoptotic cell death
Most of the apoptotic cell deaths that occur during C. elegans development occur very rapidly: the cells are generated and within ∼30 min they have been killed and turned into a cell corpse (Sulston and Horvitz 1977; Sulston et al. 1983). There is evidence in support of the notion that the transcriptional activation of egl-1 is not only necessary but also sufficient for this type of apoptotic cell deaths (referred to as “canonical” apoptotic death) and that egl-1 transcriptional upregulation marks their onset (Conradt and Horvitz 1999; Thellmann et al. 2003; Liu et al. 2006; Hatzold and Conradt 2008; Potts et al. 2009; Hirose et al. 2010; Winn et al. 2011; Hirose and Horvitz 2013; Jiang and Wu 2014; Wang et al. 2015). However, there are at least two types of apoptotic cell deaths that occur during C. elegans development for which the transcriptional upregulation of egl-1 does not seem to be sufficient: the death of the four CEMs and the death of the tail-spike cell. Furthermore, the death of the CEMs is still dependent on egl-1 function (96% of the CEMs survive in hermaphrodites lacking egl-1 function); however, the death of the tail-spike cell is only partially dependent on egl-1 (30% of the tail-spike cells survive in animals lacking egl-1 function) (Maurer et al. 2007; Nehme et al. 2010). Interestingly, rather than dying within ∼30 min, the CEMs and the tail-spike cell die ∼150 min or ∼300 min after being generated, respectively (Sulston and Horvitz 1977; Sulston et al. 1983). It has been proposed that this delay of their deaths could be the reason why egl-1 transcriptional upregulation is not sufficient for them (Nehme et al. 2010). The answer might lie in the amount of the CED-3 Caspase zymogen present in cells programmed to die. The ced-3 gene is strongly expressed in mothers of cells programmed to die and inactive CED-3 zymogens produced in the mothers are inherited to the daughters, where it presumably gets processed and activated in the daughter that is programmed to die once the onset of death has been triggered by egl-1 transcriptional upregulation (Maurer et al. 2007; Chakraborty et al. 2015). Interestingly, in both the CEMs and the tail-spike cell, transcriptional upregulation of ced-3 is observed just prior to their deaths (Maurer et al. 2007; Nehme et al. 2010). Based on these observations it has been proposed that at the time the CEMs and the tail-spike cell die, the level of the CED-3 zymogen in these cells might have decreased below a threshold (due to protein turnover) that is necessary to generate enough active CED-3 to trigger the execution phase of programmed cell death, and upregulation of ced-3 becomes necessary (Nehme et al. 2010). Based on antibody staining, CED-9 Bcl-2 and CED-4 Apaf-1 appear to be present in most if not all cells, at least during most of embryonic development (Chen et al. 2000). The transcriptional activation of ced-3 would induce new CED-3 zymogen synthesis, raising the level of the CED-3 zymogens above the necessary threshold. Interestingly, in the CEMs, the transcriptional upregulation of ced-3 occurs after the transcriptional upregulation of egl-1, indicating that the onset of CEM death is regulated by ced-3 transcriptional activation rather than egl-1 transcriptional activation (Nehme et al. 2010). In the tail-spike cell, the activation step has been modified even further: egl-1 transcriptional activation is no longer an absolute requirement; in contrast, in the presence of a functional ced-4 Apaf-1 gene, ced-3 transcriptional activation appears to be sufficient to induce the onset of tail-spike cell death (Maurer et al. 2007).
Cell Death Execution
Once the cell death program is activated, it initiates the highly regulated cell disassembly process, which includes nuclear DNA fragmentation, cytoplasm shrinkage, mitochondria elimination, and exposure of an “eat me” signal(s) such as phosphatidylserine (PS) on the surface of the dying cell to induce phagocytosis by neighboring cells or macrophages (Steller 1995). The activated caspases play crucial roles in coordinating the execution of different cell disassembly events by cleaving and activating proapoptotic protease targets and by cleaving and inactivating prosurvival protease substrates. The activated protease targets then initiate different cell killing events that contribute to the demise of the cell.
CED-3-activated cell death execution events
Nuclear DNA fragmentation:
Fragmentation of chromosomal DNA is a hallmark of apoptosis and may facilitate apoptosis by terminating DNA replication and gene transcription, which maintain the survival and the functions of the cell (Arends et al. 1990). DNA fragmentation during C. elegans apoptosis has been studied with the aid of various DNA-staining techniques, including DAPI, Feulgen (Sulston 1976), or TdT-mediated dUTP nick end labeling (TUNEL) staining (Gavrieli et al. 1992; Wu et al. 2000; Parrish et al. 2001).
So far, 11 nuclease-encoding genes have been identified to be involved in nuclear DNA degradation during apoptosis (Sulston 1976; Wu et al. 2000; Parrish et al. 2001; Wang et al. 2002; Parrish and Xue 2003; Nakagawa et al. 2010). These include nuclease defective 1 (nuc-1), CED-3 protease suppressor 6 (cps-6), cell death-related nuclease 1–7 (crn-1 to crn-7), cyclophilin 13 (cyn-13), and Dicer related 1 (dcr-1). Loss or reduction of activity in any of these genes except dcr-1 results in accumulation of TUNEL-positive cells in C. elegans embryos, suggesting that most of these nucleases are involved in resolving TUNEL-reactive DNA breaks generated during apoptosis (Sulston 1976; Wu et al. 2000; Parrish et al. 2001; Wang et al. 2002; Parrish and Xue 2003). Reduced activity in dcr-1 on its own does not show any TUNEL phenotype, but can greatly reduce the number of TUNEL-positive cells in other cell death nuclease-deficient backgrounds (Nakagawa et al. 2010), indicating that dcr-1 acts upstream of other cell death nucleases to produce TUNEL-reactive DNA breaks and likely makes the first cuts on nuclear DNA during apoptosis. Moreover, loss or reduction of activity in most of these genes (with the exception of nuc-1, crn-6, and crn-7; which encode DNase II homologs) causes delayed appearance of embryonic cell corpses or reduced cell death during embryo development and can block cell death in sensitized genetic backgrounds; suggesting that nuclear DNA degradation is important for normal progression of the apoptotic process and can promote cell killing (Parrish et al. 2001; Wang et al. 2002; Parrish and Xue 2003; Nakagawa et al. 2010). Genetic, phenotypic, and biochemical analyses indicate that these genes act sequentially in three different stages to promote DNA degradation and apoptosis (Figure 2B). First, the activated CED-3 protease cleaves the DCR-1 ribonuclease, a double-stranded RNA processing endonuclease, in the middle of the first of the 2 RNase III domains; generating a C-terminal cleavage product (tDCR-1) with one and a half RNase III domains (Nakagawa et al. 2010). tDCR-1 is capable of binding DNA and making 3′ hydroxyl DNA nicks (Nakagawa et al. 2010; Ge et al. 2014), which are labeled in the TUNEL assay. Therefore, CED-3 cleavage converts DCR-1 from an RNase to a DNase, which initiates the nuclear DNA degradation process. In the second stage, the mitochondrial endonuclease encoded by cps-6 interacting with multiple CRN nucleases, such as CRN-1, and nonnuclease factors, such as the worm apoptosis-inducing factor homolog WAH-1, to form a multi-nuclease complex (degradeosome) to catalyze stepwise DNA fragmentation; starting from turning the DNA nicks generated by tDCR-1 to single-stranded DNA gaps and double-stranded DNA breaks (Wang et al. 2002; Parrish and Xue 2003; Parrish et al. 2003). As a result, inactivation of any of the components in the degradeosome results in accumulation of TUNEL-reactive DNA ends (Wang et al. 2002; Parrish and Xue 2003). In the third stage, three DNase II homologs, NUC-1, CRN-6, and CRN-7, mediate further degradation of fragmented nuclear DNA in dying cells, with NUC-1 playing the major role in this process (Wu et al. 2000; Lai et al. 2009; Yu et al. 2015). However, the three DNase II-encoding genes do not appear to affect either the activation or progression of cell death, or the engulfment of cell corpses (Hedgecock et al. 1983; Wu et al. 2000; Parrish et al. 2001; Parrish and Xue 2003; Lai et al. 2009). They are likely involved in the cleanup step of cell death execution. In addition to its cell death function, nuc-1 is involved in degradation of DNA derived from ingested bacteria in the intestinal lumen (Sulston 1976; Wu et al. 2000).
cps-6 and wah-1 encode mitochondrial proteins which are similar to human mitochondrial endonuclease G (EndoG) and apoptosis-inducing factor (AIF), respectively (Parrish et al. 2001; Wang et al. 2002). Both EndoG and AIF have been shown to mediate nuclear DNA fragmentation in mammalian apoptosis (Susin et al. 1999; Li et al. 2001). WAH-1 can physically associate with CPS-6 and enhance the endonuclease activity of CPS-6 (Wang et al. 2002). Ectopic egl-1 expression induces the release of WAH-1 from mitochondria and its subsequent translocation to nuclei in a CED-3-dependent manner (Wang et al. 2002); suggesting that the role of mitochondria in regulating apoptosis is conserved, at least at the step of cell death execution.
PS externalization:
During apoptosis, eat me signals are expressed on the surface of the dying cell to trigger rapid clearance of the apoptotic cell (Savill et al. 1993; Savill and Fadok 2000). PS, which is normally restricted to the inner leaflet of the plasma membrane, is externalized during apoptosis and serves as an eat me signal to trigger phagocytosis (Fadok et al. 1992a; Martin et al. 1995; Fadok et al. 1998). PS exposure on the surface of apoptotic cells in C. elegans has been shown to be a conserved apoptotic event and is important for removal of apoptotic cells (Venegas and Zhou 2007; Wang et al. 2007; Zullig et al. 2007). Interestingly, both WAH-1 and its human homolog AIF, the proapoptotic factors critical for nuclear DNA degradation, are also involved in promoting PS externalization in apoptotic cells (Susin et al. 1999; Wang et al. 2007). WAH-1 accomplishes this by binding to the C. elegans phospholipid scramblase SCRM-1 located on the plasma membrane and activating its bidirectional lipid scrambling activity, leading to the exposure of PS on the surface of the dying cell. Consistently, inactivation of scrm-1 results in a mild engulfment defect and RNA interference (RNAi) knockdown of wah-1 enhances the engulfment defects of other engulfment mutants (Wang et al. 2007; Hsu and Wu 2010). Since wah-1 acts downstream of ced-3 and the release of WAH-1 from mitochondria during apoptosis is a CED-3-dependent event (Wang et al. 2002; Breckenridge et al. 2008), this represents the second CED-3-activated cell death execution event mediated by WAH-1 (Figure 2B).
Because loss of scrm-1 only causes a mild engulfment defect, other genes may contribute to PS exposure during apoptosis. Indeed, another multipass transmembrane protein CED-8 plays a more important role in externalizing PS on the surface of the apoptotic cell. ced-8 was originally identified as a gene that affects the kinetics of apoptosis and shares sequence similarity to proteins from the XK transporter family (Stanfield and Horvitz 2000). Recent studies suggest that both CED-8 and its human homolog Xkr8 are involved in PS externalization in apoptotic cells (Y. Z. Chen et al. 2013; Suzuki et al. 2013). Importantly, CED-8 is cleaved by CED-3 and this cleavage removes a short N-terminal peptide to generate a C-terminal cleavage product (acCED-8) that is both necessary and sufficient to mediate the proapoptotic and PS externalization activities of CED-8 in vivo (Y. Z. Chen et al. 2013). How acCED-8 promotes PS externalization and apoptosis is not understood. Inactivation of ced-8 enhances the cell corpse engulfment defects in animals that are deficient in either of the two major phagocytosis pathways acting in parallel in C. elegans (Reddien and Horvitz 2004; Y. Z. Chen et al. 2013; Suzuki et al. 2013), indicating that ced-8 acts through both pathways to promote phagocytosis.
Surface-exposed PS not only triggers engulfment of apoptotic cells by phagocytes, but can also lead to phagocytosis of living cells that ectopically expose PS. This occurs in animals lacking the aminophospholipid translocase transbilayer amphipath transporter 1 (TAT-1) that maintains PS asymmetry in plasma membrane (Darland-Ransom et al. 2008). These observations are confirmed by other studies in C. elegans (Wang et al. 2010; Nawa et al. 2012) and by a mammalian study in which inactivation of CDC50A, a cofactor for the human TAT-1 homolog ATP11C, causes ectopic PS exposure in living cells and their phagocytosis by macrophages (Segawa et al. 2014). The engulfment of living cells by phagocytes in the tat-1 mutants is blocked by lf mutations in PS receptor family 1 (psr-1) and ced-1, two phagocyte receptors that recognize surface-exposed PS and act in two major phagocytosis pathways (Zhou et al. 2001b; Wang et al. 2003; Darland-Ransom et al. 2008; Wang et al. 2010; Li et al. 2015; Yang et al. 2015), suggesting that externalized PS can serve as an eat me signal for both engulfment pathways.
Mitochondrial elimination:
As described above, mitochondria play an important role in regulating cell death execution in C. elegans. During apoptosis, mitochondria also undergo dramatic morphological changes, including fragmentation, reorganization of cristae structures, and increased permeability of the outer mitochondrial membrane (Jagasia et al. 2005; Cereghetti and Scorrano 2006; Parone and Martinou 2006). There are also reports that mitochondria are reduced or lost during apoptosis, which would eliminate cellular energy production and contribute to the demise of the cell (Skulachev et al. 2004; Arnoult et al. 2005). A comprehensive genetic and cell biological analysis of components of the C. elegans mitochondrial fission and fusion machinery, the dynamin GTPases DRP-1, FZO-1 (FZO mitochondrial fusion protein related), and eating defective 3 (EAT-3), indicates that defects in mitochondrial fission or fusion in C. elegans do not affect apoptosis activation (Breckenridge et al. 2008). However, loss of DRP-1 or FIS-2 (S. cerevisiae FIS1-related), a homolog of the human Fis1 fission protein, does cause a mild cell death defect that can be detected in sensitized genetic backgrounds, suggesting that fis-2 and drp-1 have minor proapoptotic roles. Genetic epistatic analysis suggests that fis-2 and drp-1 act independently of each another and downstream of ced-3 to promote apoptosis. Analysis by electron microscopy indicates that mitochondria normally reduced or eliminated in apoptotic cells persist in animals deficient in fis-2 or drp-1, indicating that DRP-1 and FIS-2 play a role in promoting mitochondrial elimination during apoptosis (Breckenridge et al. 2008). Active CED-3 protease can cleave DRP-1 in vitro and this cleavage is critical for DRP-1’s proapoptotic function in vivo, but dispensable for its function in mitochondrial fission (Breckenridge et al. 2008). Furthermore, the C-terminal cleavage product of DRP-1 appears to be important for activating DRP-1’s proapoptotic function, together with the full-length DRP-1 protein. Therefore, fis-2 and drp-1 represent two novel cell death execution pathways acting downstream of ced-3 to promote mitochondrial elimination (Figure 2B).
Inactivation of survival signals:
In living cells, multiple cell death inhibitors or survival factors work together to maintain the viability and functions of the cell. During apoptosis, these survival factors are inactivated to allow apoptosis to proceed (Danial and Korsmeyer 2004). In C. elegans, the key cell death inhibitor CED-9 is an excellent substrate of CED-3 in vitro. Because the two cleavage products of CED-9 generated by CED-3 cleavage display significantly weaker death protective activity (Xue and Horvitz 1997), CED-3 cleavage of CED-9 could markedly compromise its cell death inhibitory activity.
Another well-known cell survival pathway is the phosphoinositide 3-kinase (PI3K)/AKT signaling pathway that promotes cell growth, proliferation, and survival in diverse organisms (Luo et al. 2003; Cully et al. 2006). How this crucial survival pathway is inactivated to promote apoptosis is not well understood. From a CED-3 protease suppressor screen, a CED-3 substrate, CNT-1, was identified and found to act downstream of CED-3 to promote apoptosis (Nakagawa et al. 2014). CNT-1 is cleaved during apoptosis to generate an N-terminal phosphoinositide (PI)-binding cleavage product, tCNT-1. tCNT-1 then translocates from the cytoplasm to the plasma membrane to block AKT binding to phosphatidylinositol (3,4,5)-trisphosphate (PIP3), thereby inhibiting AKT activation and its prosurvival activity (Nakagawa et al. 2014). CNT-1 defines a novel, caspase-activated negative regulator of the AKT survival pathway.
There are probably additional CED-3 substrates that are important for other aspects of cell death execution, such as cytoplasm shrinkage, nuclear membrane breakdown, and cell corpse engulfment. Molecular genetic characterization of additional CED-3 protease suppressors should lead to identification of additional CED-3 substrates and CED-3-activated cell death execution events.
Clearance of Apoptotic Cells
When a cell undergoes apoptosis, eat me signals are rapidly exposed on the surface of the apoptotic cell (Fadok et al. 2001). These signals are recognized by receptors on the engulfing cells to trigger the phagocytosis of apoptotic cells (reviewed by Hochreiter-Hufford and Ravichandran 2013). The engulfment process includes membrane extension and cytoskeleton rearrangement of growing pseudopods around an apoptotic cell, and the enclosure of the pseudopods to form a phagosome.
Unlike flies or humans, C. elegans does not have “professional” phagocytes, such as mobile macrophages; rather, apoptotic cells are engulfed by their neighboring cells. Cell types such as hypodermal cells (which constitute the external epithelium), muscle cells, and intestinal cells have been shown to function as engulfing cells to remove somatic apoptotic cells (Robertson and Thomson 1982; Sulston et al. 1983; Zhou et al. 2001b; Hsieh et al. 2012). Germ cell corpses are specifically engulfed by gonadal sheath cells, which wrap around the germ line syncytium (Gumienny et al. 2001).
Presentation of eat me signals
Thus far, the best known eat me signal on apoptotic cells is PS, which is externalized from the cytosolic (inner) leaflet to the noncytosolic (outer) leaflet of the plasma membrane during apoptosis (Fadok et al. 1992b; Fadeel and Xue 2009). A common feature of all eukaryotic membranes is the asymmetric distribution of different phospholipids in the lipid bilayer. For example, aminophospholipids, phosphatidylethanolamine, and PS are restricted to the inner leaflet of the plasma membrane in living cells. Externalization of PS on the cell surface is a hallmark of apoptosis; exposed PS is a conserved eat me signal that triggers phagocytosis in many organisms, including C. elegans (Fadeel and Xue 2009). Using a secreted PS-binding protein GFP fusion such as the Annexin V::GFP fusion (sAnxV::GFP), the MFG-E8::GFP, or the secreted GFP::lactadherin fusion (sGFP::LactC1C2) as a PS sensor; exposed PS is detected on the surface of apoptotic cells in C. elegans (Fadok et al. 2001; Venegas and Zhou 2007; Wang et al. 2007; Zullig et al. 2007; Mapes et al. 2012; Zhang et al. 2012).
Two bidirectional phospholipid scramblases, SCRM-1 and SCRM-3, have been implicated in PS exposure on the surface of apoptotic cells (Venegas and Zhou 2007; Wang et al. 2007). Loss of scrm-1 or scrm-3 (also called plsc-1), which encodes two of the eight C. elegans phospholipid scramblases, results in reduced PS exposure on the surface of apoptotic germ cells and a defect in the removal of apoptotic cells (Venegas and Zhou 2007; Wang et al. 2007; Hsu and Wu 2010). Loss of scrm-1 or scrm-3 only partially reduces PS exposure in apoptotic cells, suggesting that additional factors or lipid transporters are involved in mediating PS exposure in apoptotic cells. As discussed above, the mitochondrial apoptogenic factor WAH-1 also affects PS externalization during apoptosis (Wang et al. 2007). WAH-1 promotes PS externalization by binding to SCRM-1 and activating the phospholipid scrambling activity of SCRM-1 (Figure 2B). The CED-8 protein, a homolog of the XK family transporters, is critical for mediating PS externalization in somatic apoptotic cells (Y. Z. Chen et al. 2013; Suzuki et al. 2013). Cleavage of CED-8 by CED-3 during apoptosis generates a C-terminal cleavage product, acCED-8, that promotes PS externalization in apoptotic cells and is sufficient to induce ectopic PS externalization in living cells (Y. Z. Chen et al. 2013). The ABC transporter CED-7 has been proposed to mediate PS exposure in somatic apoptotic cells using the MFG-E8::GFP PS sensor (Venegas and Zhou 2007). However, multiple studies using other PS sensors show that CED-7 does not promote PS externalization in apoptotic cells (Zullig et al. 2007; Mapes et al. 2012; Zhang et al. 2012), and instead, plays a role in the efflux of PS from apoptotic cells (Mapes et al. 2012; Zhang et al. 2012).
Surface PS expression on phagocytes
Interestingly, PS exposure was detected not only on the membranes of apoptotic cells but also on those of engulfing cells (Mapes et al. 2012; Zhang et al. 2012). Externalized PS appears early on the surface of the dying cells and decreases in older or unengulfed apoptotic cells. This decrease in surface PS exposure depends on a secreted extracellular protein transthyretin-related family domain 52 (TTR-52) and CED-7, a homolog of the mammalian ABC1 transporters (Mapes et al. 2012; Zhang et al. 2012). TTR-52 is expressed in and secreted from the endoderm (Wang et al. 2010), while CED-7 is widely expressed and localized on the surface of somatic cells (Wu and Horvitz 1998a). TTR-52 and CED-7 together with CED-1, an engulfment receptor localized on the surface of engulfing cells (Zhou et al. 2001b), are required for PS appearance on the surface of the phagocytes (Mapes et al. 2012; Zhang et al. 2012). Immunoelectron microscopy analysis of embryos expressing sAnxV::GFP reveals the presence of extracellular PS-containing vesicles between the dying cells and their neighboring cells in a ced-7- and ttr-52-dependent manner. It has been proposed that CED-7 and TTR-52 promote the efflux of PS from apoptotic cells by generating extracellular PS vesicles, which cause PS appearance on the surface of phagocytes through CED-1 (Mapes et al. 2012). Moreover, sGFP::LactC1C2, which labels apoptotic cells but not phagocytes, prevents sAnxV::GFP from labeling phagocytes and compromises phagocytosis (Mapes et al. 2012). Therefore, PS expression on the phagocytes is also important for the engulfment of apoptotic cells.
Nose resistant to fluoxetine 5 (NRF-5), a secreted lipid transfer/LPS-binding family protein, is also important for PS appearance on the surface of phagocytes (Zhang et al. 2012). NRF-5 binds TTR-52 and PS, and displays a lipid transfer activity in vitro. NRF-5 may act with TTR-52 and CED-7 to mediate PS transfer from apoptotic cells to engulfing cells. How PS expression on phagocytes facilitates apoptotic cell clearance is not clear. One possibility is that appearance of PS on the surface of engulfing cells may alter the activity of membrane proteins that are important for the removal of cell corpses, and thus promote the engulfment process. Alternatively, PS may act as a homotypic ligand to tether the apoptotic cell to the engulfing cell through a bipartite PS-binding bridging molecule and thus facilitates the engulfment process (Mapes et al. 2012).
Engulfment receptors and signaling pathways
Forward and reverse genetics have identified ∼20 genes required for the engulfment of apoptotic cells. To assess whether these genes act in the same or separate pathways during the engulfment process, genetic analyses have been performed to position two genes at a time. Double mutants of genes acting in different pathways have a stronger engulfment defect (e.g., more persistent cell corpse numbers or longer duration of cell corpses) than those of single mutants alone or double mutants of genes acting in the same pathway (Ellis et al. 1991). On the basis of such analyses, three partially redundant pathways have been established that mediate the engulfment process.
The CED-1, CED-6, and CED-7 pathway:
This pathway comprises the engulfment receptor CED-1, a homolog of the mammalian MEGF10 protein (Hamon et al. 2006). The ced-1 gene is expressed and functions in engulfing cells, but not in apoptotic cells, during cell corpse engulfment (Zhou et al. 2001b). The CED-1::GFP fusion was found to cluster around apoptotic cells, and this clustering completely depends on CED-7 (Zhou et al. 2001b) and partially depends on the secreted extracellular protein TTR-52 (Wang et al. 2010). Therefore, the recognition and binding of apoptotic cells by CED-1 requires TTR-52 and CED-7. TTR-52 binds both PS and the extracellular domain of CED-1 in vitro and likely functions as a bridging molecule that mediates recognition of apoptotic cells by cross-linking the exposed PS eat me signal with the engulfment receptor CED-1 (Figure 2B). In addition, a recent study suggests that the extracellular region of CED-1 could directly bind PS in vitro, when fused to GST (Li et al. 2015).
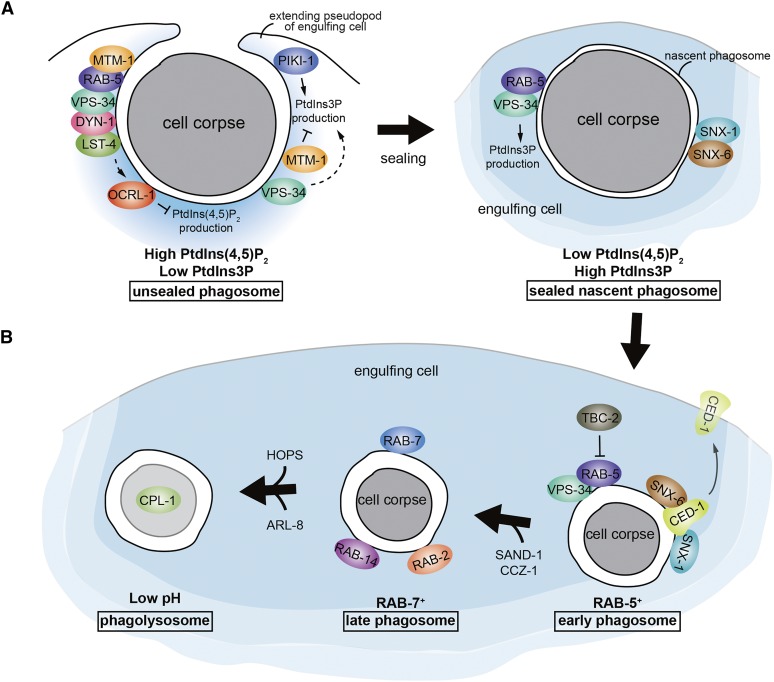
Upon binding to an apoptotic cell through TTR-52 or directly, CED-1 transduces the engulfment signal via the adaptor protein CED-6 (homologous to the mammalian GULP protein) and DYN-1, a member of the large GTPase family that regulate vesicle transport events (Clark et al. 1997), to promote the internalization and subsequent degradation of apoptotic cells (Liu and Hengartner 1999; Yu et al. 2006; Guo et al. 2010; Wang et al. 2010). CED-6 contains a phosphotyrosine binding domain (Liu and Hengartner 1998) and may directly bind to the intracellular domain of CED-1 (Su et al. 2002). DYN-1 clusters on pseudopods around an apoptotic cell in a ced-1-, ced-6-, and ced-7-dependent manner (Yu et al. 2006). Therefore, CED-6 may link CED-1 signaling to DYN-1. Using the endosomal marker HGRS-1::GFP to track endosomes during the engulfment process, Yu et al. (2006) found that endosomes, which displayed a punctate localization pattern in the cytoplasm, were gradually recruited to phagocytic cups and phagosomes around apoptotic cells. The incorporation of endosome vesicles to phagocytic membranes requires CED-1 and DYN-1. The role of dyn-1 in internalization of cell corpses is controversial. A parallel study by Kinchen et al. (2008) showed that clustering of DYN-1 around germ cell corpses was significantly reduced not only in ced-1, ced-6, or ced-7 mutants, but also in ced-5, ced-10, or ced-12 mutants (see below) defective in cell corpse engulfment; suggesting that DYN-1 is recruited at a stage following corpse recognition and internalization. Consistently, it has been shown that DYN-1 acts at an early stage in phagosome maturation (Kinchen et al. 2008; Almendinger et al. 2011; D. Chen et al. 2013; Cheng et al. 2015), particularly in phagosome sealing (see below) (Cheng et al. 2015). However, Yu et al. (2006) observed DYN-1 clustering around germ cell corpses in ced-5, ced-10, or ced-12 mutants and their ultrastructural studies showed that some germ cell corpses of dyn-1 mutants were either not internalized or internalized but not degraded, indicating that dyn-1 is important for both internalization and degradation of germ cell corpses.
DYN-1 also promotes actin assembly at the phagocytic cup during the engulfment of both embryonic and germ cell corpses, probably through different mechanisms (D. Chen et al. 2013; Shen et al. 2013). During the engulfment of embryonic cell corpses, clathrin heavy chain 1 (CHC-1) and clathrin adaptor protein epsin 1 (EPN-1), but not adaptor protein AP2, are recruited to phagocytic cups (Shen et al. 2013). This recruitment requires DYN-1, suggesting that dyn-1 acts upstream of epn-1 and chc-1 during this process. Inactivating chc-1 or epn-1 impairs F-actin polymerization and stability underneath the phagocytic cup, similar to those observed in mutants defective in ced-1, ced-6, or dyn-1, including the repeated retraction of actin filaments in the phagocytic cups and the loss of the engulfing-dying cell adhesion (Shen et al. 2013). During the clearance of germ cell corpses, clathrin and AP2 are recruited to phagocytic cups in a mutually-dependent manner (D. Chen et al. 2013; Shen et al. 2013). Genetic analysis revealed that chc-1 and apb-1 (AP2 subunit) act downstream of ced-1 and ced-6, but upstream of dyn-1 in this process (D. Chen et al. 2013). Consistently, loss of chc-1 or apb-1 inhibits DYN-1 recruitment to phagosomes. The CED-1 receptor interacts with the α subunit of AP2, while the CED-6/Gulp adaptor forms a complex with both CHC-1 and the AP2 complex (D. Chen et al. 2013). In addition, DYN-1, clathrin, and AP2 are reported to facilitate the maturation of apoptotic cell-containing phagosomes necessary for corpse degradation in adult germline (see below) (Kinchen et al. 2008; Almendinger et al. 2011; D. Chen et al. 2013; Cheng et al. 2015). However, Shen et al. (2013) observed no significant accumulation of germ cell corpses when AP2 was inactivated, and found no involvement of clathrin in maturation of phagosomes containing embryonic cell corpses. The discrepancies could be due to differences in RNAi efficiency in AP2 inactivation and a unique role of clathrin in the maturation of phagosomes containing germ cell corpses, but not embryonic cell corpses.
Both small GTPases CED-10 (Rac) and CDC-42 (Cdc42) have been implicated to act downstream of CED-1 and CED-6 to promote the cytoskeleton rearrangement in engulfing cells (Kinchen et al. 2005; Neukomm et al. 2014); however, this has been debated. For example, the recruitment of EPN-1 and CHC-1 to phagocytic cups during cell corpse engulfment is independent of CED-5, CED-12, or CED-10; suggesting that the remodeling of actin regulated by the CED-1-EPN-1-clathrin pathway may not require CED-10 (Shen et al. 2013). Instead, ced-10 and cdc-42 have been placed in two separate pathways, both of which act in parallel to that of ced-1, ced-6, and ced-7 (see below) (Ellis et al. 1991; Reddien and Horvitz 2000; Yu et al. 2006; Hsieh et al. 2012; Shen et al. 2013).
The CED-5 and CED-12 pathway:
The second signaling pathway of cell corpse engulfment is mediated by at least three receptors: PSR-1 (Wang et al. 2003); more of MS 5 (MOM-5), which encodes a C. elegans Frizzled homolog (Cabello et al. 2010); and integrin α 1/paralyzed arrest at twofold; integrin β subunit 3 (INA-1/PAT-3) (Figure 2B) (Hsu and Wu 2010). MOM-5 appears to be a predominant engulfment receptor for removal of embryonic cell corpses in this signaling pathway, as the mom-5 null mutants have a similar extent of engulfment defect as ced-5 null mutants during early embryogenesis (Cabello et al. 2010), whereas the engulfment defects of psr-1, ina-1, or pat-3 mutants are much weaker than those of ced-5 and mom-5 (Cabello et al. 2010; Hsu and Wu 2010). Although how MOM-5 may recognize apoptotic cells is not yet clear, the extracellular domains of PSR-1 and INA-1 likely bind to exposed PS on the surface of apoptotic cells, and both PSR-1 and INA-1 are found to cluster on the surface of apoptotic cells (Wang et al. 2003; Hsu and Wu 2010; Yang et al. 2015). A lysine-rich motif in the extracellular domain of PSR-1 is critical for PS binding and PS-induced oligomerization of PSR-1 in vitro, suggesting a mechanism by which PSR-1 activates the cell corpse engulfment process (Yang et al. 2015). Interestingly, in addition to mediating recognition and clearance of apoptotic cells with surface-exposed PS, PSR-1 plays a critical role in promoting axonal fusion during regeneration of neurons with severed axons by mediating recognition of the PS “save me” signal exposed on the surface of the severed distal axon fragment (Neumann et al. 2015). Therefore, PSR-1 is a PS receptor for both apoptotic and nonapoptotic events in C. elegans. In mammals, the PSR-1 homolog, PSR, has been reported to have both apoptotic and nonapoptotic roles during animal development and tissue homeostasis (Fadok et al. 2000; Hisatomi et al. 2003; Li et al. 2003; Hong et al. 2004; Kunisaki et al. 2004; Zakharova et al. 2009), but there are conflicting reports on whether it has a detectable role in clearance of apoptotic cells (Bose et al. 2004; Mitchell et al. 2006). This is probably due to different detection methods and the presence of multiple independent phagocytosis pathways that are even more complex than those in C. elegans (Savill and Fadok 2000).
All three receptors, MOM-5, PSR-1, and INA-1/PAT-3, signal through the bipartite complex CED-5 (DOCK180)/CED-12 (ELMO) to regulate the activation of CED-10 (RAC1) GTPase (Wu and Horvitz 1998b; Reddien and Horvitz 2000; Gumienny et al. 2001; Wu et al. 2001; Zhou et al. 2001a; Brugnera et al. 2002; Wang et al. 2003; Yang et al. 2015). The CED-10 GTPase cycles between GTP-bound (“on”) and GDP-bound (“off”) states, in which GTP loading of GTPases is promoted by GEFs. Studies in mammalian models showed that both DOCK180 and ELMO contact and mediate the exchange of GDP and GTP on RAC1 (Brugnera et al. 2002; Lu et al. 2004, 2005). It is possible that their C. elegans homologs, CED-5 and CED-12, also have a GEF activity toward CED-10. Activation of CED-10 may subsequently lead to actin rearrangement required for the pseudopod extension around an apoptotic cell (Kinchen et al. 2005). Nucleoside diphosphate kinase 1 (NDK-1), similar to NM23-H1/H2, which has NDK activity, is reported to act downstream of CED-10 to promote cell corpse engulfment, but the molecular basis of NKD-1 function is still unknown (Fancsalszky et al. 2014).
The three phagocyte receptors, PSR-1, MOM-5, and INA-1/PAT-3, activate the CED-5-CED-12-CED-10 signaling pathways in different ways (Figure 2B). The intracellular region of PSR-1 directly interacts with CED-5 and CED-12 (Wang et al. 2003), whereas MOM-5 and INA-1 connect to the CED-5-CED-12-CED-10 signaling pathway via the adaptor protein CED-2(CRKII) (Cabello et al. 2010; Hsu and Wu 2010). Biochemical and crystal structural analyses revealed that CED-2 interacts with CED-5 through the SRC homology 3 domain of CED-2 and the N-terminal region of CED-5 (Kang et al. 2011). MOM-5 regulates CED-2 through atypical Wnt signaling that includes GSK-3 kinase (GSK3β) and APR-1 (APC) (Cabello et al. 2010). INA-1 signals through a nonreceptor tyrosine kinase SRC-1 (SRC), which links the INA-1 intracellular domain to CED-2 (Hsu and Wu 2010). In addition to these three receptors, the UNC-73 (Trio homolog with a GEF activity)-abnormal cell migration, RhoG homolog 2 (MIG-2) signaling module activates CED-10, likely through CED-5 and CED-12 (deBakker et al. 2004). However, the receptor acting upstream of UNC-73-MIG-2 is unknown. Interestingly, all genes in this pathway, except psr-1, also affect the migration of the gonadal distal tip cells (DTCs) in hermaphrodites, suggesting the existence of a conserved-motility machinery used in the membrane extension process during both cell corpse engulfment and DTC migration (Wu and Horvitz 1998b; Reddien and Horvitz 2000; Gumienny et al. 2001; Wu et al. 2001; Zhou et al. 2001a; Brugnera et al. 2002; Cabello et al. 2010; Hsu and Wu 2010).
The PAT-2 and PAT-3 pathway:
The third engulfment pathway is mediated by the integrin PAT-2/PAT-3 (Hsieh et al. 2012; Neukomm et al. 2014). The integrin α subunit PAT-2, like the other integrin α subunit INA-1, clusters on the surface of apoptotic cells (Hsu and Wu 2010; Hsieh et al. 2012), but whether PAT-2 may bind to exposed PS is not known. PAT-2 signaling leads to the recruitment of the GEF protein UIG-1, likely via the UIG-1-interacting protein UNC-112 and UIG-1’s target CDC-42 GTPase, to the phagocytic cups and the subsequent activation of CDC-42 GTPase (Figure 2B) (Hsieh et al. 2012; Neukomm et al. 2014). In addition to the PAT-2 pathway, the CED-1, CED-6, and CED-7 pathway may also act upstream of CDC-42 (Neukomm et al. 2014). PAT-2 functions in muscle cells for apoptotic cell engulfment. By contrast, INA-1 and the engulfment receptor CED-1 preferentially act in epithelial cells to mediate cell corpse removal (Hsieh et al. 2012). Therefore, different types of engulfing cells appear to use distinct repertoires of phagocyte receptors at the whole organism level.
Negative regulators of the engulfment process
MTM-1, SRGP-1, and PDR-1:
The CED-5-CED-12 signaling pathway is negatively regulated by the PI phosphatase protein MTM-1 (Zou et al. 2009; Neukomm et al. 2011b), GAP SRGP-1 (Neukomm et al. 2011a), and the E3 ubiquitin ligase PDR-1 in engulfing cells. Loss of mtm-1, srgp-1, or pdr-1 increases the kinetics of embryonic cell corpse removal, whereas overexpression of any of these genes results in accumulation of embryonic cell corpses. MTM-1, similar to its human homolog myotubularin 1, has phosphatase activity toward both phosphatidylinositol-3-phosphate [PtdIns(3)P] and phosphatidylinositol-3,5-bisphosphate [PtdIns(3,5)P2], and the phosphatase activity is required for its negative regulatory function for cell corpse engulfment (Neukomm et al. 2011b; Lu et al. 2012). The CED-12 pleckstrin homology domain can bind to PtdIns(3,5)P2, suggesting that MTM-1 may inhibit the recruitment of CED-12 to the plasma membrane by dephosphorylating PtdIns(3,5)P2 and thus reduces the CED-10 activity (Neukomm et al. 2011b). In addition, MTM-1 also plays a role in degradation of germ cell corpses (see below).
The other two negative regulators in the CED-5-CED-12 signaling pathway, SRGP-1 and PDR-1, reduce the activity and the protein level of CED-10 GTPase, respectively (Neukomm et al. 2011a; Cabello et al. 2014). PDR-1 is the C. elegans homolog of human E3 ubiquitin ligase Parkin, whose mutations are considered causative of autosomal recessive juvenile parkinsonism (Kitada et al. 1998). The E3 ubiquitin ligases constitute the third step of the ubiquitination process and have a role in catalyzing the conjugation of ubiquitin to a lysine residue in the target proteins for proteasome-mediated degradation. PDR-1 binds to CED-10 and promotes poly-ubiquitination and subsequent proteasomal degradation of CED-10 (Cabello et al. 2014). The GAP protein SRGP-1 binds to CED-10 and promotes CED-10 GTP hydrolysis in vitro, and thereby negatively regulates CED-10 GTPase activity (Neukomm et al. 2011a). Recent findings have revealed that SRGP-1 also has a GAP activity toward CDC-42 in vitro and that loss of srgp-1 suppresses the engulfment defect of animals treated with cdc-42(RNAi), suggesting that srgp-1 also negatively regulates the cdc-42-mediated engulfment pathway during development (Neukomm et al. 2014).
Interestingly, enhanced engulfment due to loss of mtm-1 or srgp-1 leads to increased elimination of sick cells, such as cells on the verge of death in weak ced-3 mutants with limited caspase activity or nonapoptotic, cytotoxic cell death induced by osmotic imbalance (Neukomm et al. 2011a,b). In contrast, loss of engulfment activity could promote cell survival (see below) (Hoeppner et al. 2001; Reddien et al. 2001; Chakraborty et al. 2015). Therefore, the engulfment pathway might be used generally to identify and eliminate not only apoptotic but also sick or damaged cells (Neukomm et al. 2011a). It is proposed that the sick or damaged cells can be either tolerated or eaten alive by an engulfing cell, depending on the strength of the engulfment signal sent from the sick or damaged cell and the engulfment machinery of its neighboring cells (Neukomm et al. 2011a).
PGRN-1:
PGRN-1 is the C. elegans homolog of progranulin, which is a highly conserved secretory glycoprotein and can be cleaved proteolytically into 6-kDa cysteine-rich granulins (Bateman and Bennett 2009). PGRN-1 is expressed in and secreted from neurons and intestine (Kao et al. 2011). Loss of pgrn-1 enhanced kinetics of apoptotic cell removal. Similarly, loss of progranulin accelerates phagocytosis of apoptotic thymocytes by mouse macrophages (Kao et al. 2011). These findings and the fact that mutations in human progranulin are the major cause of familial frontotemporal lobar degeneration (Baker et al. 2006; Cruts et al. 2006) raise a possibility that inadequate progranulin levels may promote death of cells, such as neurons, to result in neurodegeneration by disrupting the kinetics of cell corpse clearance (Kao et al. 2011). How PGRN-1 or progranulin regulates the rate of cell corpse removal is not clear.
ABL-1, SLI-1, and SWAN-1:
Three genes, abl-1 (related to oncogene ABL), suppressor of lineage defect 1 (sli-1), and seven WD repeats, AN11 family 1 (swan-1), negatively regulate the process of cell corpse engulfment independently of the aforementioned three signaling pathways (Yang et al. 2006; Hurwitz et al. 2009; Anderson et al. 2012). These three genes all function in engulfing cells, but not in apoptotic cells. The cytoskeletal regulator ABL-1 (Abl) kinase, opposes the engulfment of apoptotic cells by inhibiting Abl-interacting protein 1 (ABI-1) (Hurwitz et al. 2009). SLI-1, an E3 ubiquitin ligase and adaptor protein, inhibits the engulfment process in parallel to ABL-1 via a ligase-independent, yet-uncharacterized mechanism (Anderson et al. 2012). SWAN-1 likely inhibits the engulfment process by acting as an inhibitor of CED-10 GTPase (Yang et al. 2006). However, unlike srgp-1, pgrn-1, or mtm-1; inactivation of abl-1, sli-1, or swan-1 does not increase the rate of cell corpse clearance (Yang et al. 2006; Hurwitz et al. 2009; Anderson et al. 2012).
Engulfment promotes apoptosis
The engulfment process not only passively removes cell corpses but also actively kills cells. Mutations that partially inactivate killer genes, such as egl-1, ced-3, and ced-4, allow the survival of some cells that are programmed to die. Interestingly, mutations in ced-1, ced-2, ced-5, ced-6, ced-7, ced-10, or ced-12 that block the engulfment process, enhance the frequency of cell survival in the mutants with compromised killer genes (Hoeppner et al. 2001; Reddien et al. 2001). Furthermore, blocking engulfment is sufficient to cause the survival and differentiation of some cells that would normally die, as observed using a specific differentiated cell marker (Reddien et al. 2001). A microscopy analysis reveals that these cells begin the dying process (based on morphological appearance) normally, but eventually recover and survive (Hoeppner et al. 2001; Reddien et al. 2001). These results support a model that engulfing cells function to ensure that cells fated to die would die irreversibly, after the initial stages of cell death activation.
By following the division of a specific cell (NSM mother) along the dorsoventral axis that generates a ventral surviving daughter (NSM) and a dorsal apoptotic daughter, Chakraborty et al. (2015) found that the engulfment receptor CED-1 is required for the formation of a dorsoventral gradient of CED-3 caspase activity in the mother and the enhanced level of CED-3 protein in the apoptotic daughter (Chakraborty et al. 2015). CED-1 is preferentially localized to the cell surface of neighboring cells adjacent to the dorsal side of the mother. These data together support a model that CED-1, and likely other components of the engulfment pathways, may promote apoptosis by inducing asymmetric localization of apoptotic factors in mothers of cells destined to die and the unequal segregation of apoptotic potential into apoptotic and surviving daughters.
Formation and maturation of phagosomes
The internalization of an apoptotic cell by a phagocytic cell results in the formation and eventual sealing of a phagosome (membrane-bound compartments containing the phagocytosed target), in which the internalized cell corpse is rapidly degraded (Lu and Zhou 2012; Wang and Yang 2016). This internal degradation may avoid inflammatory and autoimmune responses caused by the release of potentially harmful contents of apoptotic cells. The nascent phagosome-containing apoptotic cells undergo sequential fusion with early (sorting) endosomes, late endosomes, and eventually lysosomes, as well as the gradual acidification of the phagosomal lumen; a process called phagosome maturation (Lu and Zhou 2012; Wang and Yang 2016). The maturation process enables nascent phagosomes to gradually acquire the properties of the donor organelles, including the specific membrane markers, lumen contents, and progressive acidification. Sequential fusion with these organelles results in the gradual conversion of the nascent phagosome into a phagolysosome, which, like a lysosome, contains various digestive enzymes and provides an optimal highly acidic environment (pH ≤ 5.0) for the degradative enzymes to function (Vieira et al. 2002).
Studies using genetic, biochemical, and cell biology tools have revealed the importance of PtdIns(3)P and its kinase and phosphatase in sealing of phagosomes. In addition, multiple Rab small GTPases and their regulators, and the lipid second messenger PtdIns(3)P and its effectors have been shown to facilitate phagosome maturation. Receptor CED-1 and dynamin DYN-1 act through these molecules to promote phagosome maturation (see below). Schematic drawing of sequential steps during formation and maturation of phagosomes and key proteins involved in each step are shown in Figure 4.
Figure 4.
The formation and maturation of a phagosome containing a cell corpse. Key proteins that act during (A) formation and (B) maturation of a phagosome are shown. Arrows and block arrows indicate activation or inhibition, respectively, and the dashed line indicates a proposed function.
Sealing of phagosomes:
PIs are lipid signaling molecules important for multiple intracellular membrane trafficking events (Di Paolo and De Camilli 2006; Juhasz et al. 2008). During the removal of apoptotic cells in C. elegans, phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] and PtdIns(3)P accumulate transiently on unsealed and fully-sealed phagosomes, respectively, and are both involved in phagosome sealing (Yu et al. 2008; Lu et al. 2012; Shen et al. 2013; Cheng et al. 2015). PtdIns(4,5)P2 and PtdIns(3)P are key determinants of phagosome formation and maturation in mammalian cultured cells, respectively (Bohdanowicz and Grinstein 2013). Two PI3 kinases, phosphoinositide-3 kinase 1 (PIKI-1) and VPS-34 (related to yeast vacuolar protein sorting factor), which catalyze the production of PtdIns(3)P, coordinate with MTM-1 to regulate PtdIns3P levels on unsealed phagosomes (Figure 4A) (Lu et al. 2012; Cheng et al. 2015). Phagosomal localization of PIKI-1 and VPS-34 requires DYN-1, and DYN-1 directly interacts with VPS-34 in vitro (Kinchen et al. 2008; Lu et al. 2012). PIKI-1 associates with extending pseudopods and nascent phagosomes where it functions with MTM-1 to regulate PtdIns3P levels for phagosomal sealing (Cheng et al. 2015). VPS-34 contributes to the PtdIns3P production at the sealing stage and on sealed phagosomes (Cheng et al. 2015). Cheng et al. (2015) proposed a coincident detection mechanism that controls phagosome sealing and couples sealing with the switch of membrane identity from PtdIns(4,5)P2-enriched, unsealed phagosomes to PtdIns(3)P-enriched fully-sealed phagosomes. The coincident detection code, consisting of PtdIns(4,5)P2, PtdIns(3)P, and MTM-1, recruits lateral signaling target 4 (LST-4) and subsequently DYN-1 to unsealed phagosomes to regulate sealing; probably in a similar way as the scission of endocytic vesicles (Figure 4A) (Lundmark and Carlsson 2009; Lu et al. 2012; Y. Z. Chen et al. 2013; Cheng et al. 2015). In addition, sealing of phagosomes requires timely depletion of PtdIns(4,5)P2 by inositol-5-phosphatase OCRL-1 (Lowe’s oculocerebrorenal syndrome protein) (Bohdanowicz et al. 2012; Cheng et al. 2015). It is not yet clear how OCRL-1 is regulated when phagosomes are being sealed.
Beyond the sealing process, roles and localization of PIKI-1 and MTM-1 appear different during degradation of germ cell corpses (Cheng et al. 2015) and some somatic cells (Kinchen et al. 2008; Lu et al. 2012). In adult germline, timely depletion of PtdIns(4,5)P2 may release MTM-1, LST-4, DYN-1, and therefore PIKI-1, from phagosomes. This completes the sealing process and allows subsequent accumulation of PtdIns(3)P on fully-sealed phagosomes (Bohdanowicz et al. 2012; Cheng et al. 2015), and VPS-34 is thus the predominant player in generating PtdIns3P on sealed phagosomes during subsequent phagosome maturation (Cheng et al. 2015). In contrast, in embryos, PIKI-1 and MTM-1 have been reported to act with VPS-34 in the regulation of PtdIns(3)P dynamics during maturation of phagosomes containing specific apoptotic cells (Kinchen et al. 2008; Lu et al. 2012).
Rab small GTPases in phagosome maturation:
Rab proteins are small GTPases that cycle between membrane-bound and cytosolic states in a nucleotide-dependent manner. Membrane-bound, GTP-bound Rab proteins act as tethering factors to promote organelle-organelle or organelle-membrane fusion. RAB-5 is detected on the phagosomal membrane once the nascent phagosome is formed during cell corpse clearance (He et al. 2010). The recruitment of RAB-5 and its effector VPS-34 to phagosomes is mutually dependent and DYN-1-dependent, suggesting a model in which DYN-1 acts upstream of RAB-5 and VPS-34, while RAB-5 and VPS-34 may act as a positive feedback system (Figure 4A) (Kinchen et al. 2008; He et al. 2009). The finding that mammalian Vps34 may act as a bridging molecule to connect Dyn2 and Rab5 in cultured cells suggests that the same protein-protein interaction between DYN-1, VPS-34, and RAB-5 might occur in C. elegans (Kinchen et al. 2008). Thus, RAB-5 might affect the recruitment of VPS-34, which, in turn, might contribute to further recruitment of RAB-5 to the phagosomes.
Following RAB-5 recruitment, three other small GTPases, RAB-2, RAB-14, and RAB-7, are sequentially recruited to the phagosome containing an apoptotic cell (Figure 4) (Mangahas et al. 2008; Yu et al. 2008; Guo et al. 2010). Like RAB-5, RAB-2 and RAB-14 show transient recruitment to phagosomes, whereas RAB-7 persists until the engulfed cell corpse is degraded (Mangahas et al. 2008; Yu et al. 2008; Guo et al. 2010; He et al. 2010). The rab-2; rab-14 double mutant contains more persistent cell corpses than either single mutant, suggesting that RAB-2 and RAB-14 act redundantly to degrade apoptotic cells (Guo et al. 2010). Examination of the functions of RAB-2, RAB-14, and RAB-7 in the formation of phagolysosomes suggests that RAB-2 and RAB-14 play partially-redundant roles in recruiting and tethering lysosomes to phagosomes, whereas RAB-7 acts subsequently to promote the fusion of these organelles (Guo et al. 2010). In addition, RAB-2 and RAB-14 cause acidification of the phagosomal lumen in a partially-redundant manner, while RAB-7 does not (Lu et al. 2008; Mangahas et al. 2008; Guo et al. 2010).
The binding of RAB-5 to phagosomes is transient, as RAB-5 rapidly dissociates from phagosomes (Kitano et al. 2008; He et al. 2010). The dissociation process requires TBC-2, a Rab GAP (Li et al. 2009; Chotard et al. 2010). Loss of tbc-2 and/or overexpression of RAB-5(Q78L), the constitutive active form of RAB-5, delays the release of RAB-5 from phagosomes and subsequent events of phagosome maturation, including PtdIns(3)P dynamics, the recruitment of RAB-7, the formation of phagolysosomes, and the acidification of phagosomal lumen; manifesting the importance of RAB-5 dissociation from phagosomes (Li et al. 2009).
The transition of a phagosome from the RAB-5-positive early stage to the RAB-7-positive late stage requires a protein complex composed of SAND endocytosis protein family 1 (SAND-1) and CCZ-1, the C. elegans homologs of yeast and mammalian Mon1 and ccz1, respectively (Kinchen and Ravichandran 2010; Nieto et al. 2010). In sand-1 or ccz-1 mutants the apoptotic cell-containing phagosomes are arrested at the RAB-5-positive and RAB-7-negative stage (Kinchen and Ravichandran 2010; Nieto et al. 2010), suggesting that SAND-1 and CCZ-1 are necessary for phagosomes to release RAB-5 and to acquire RAB-7. The mammalian Mon1 interacts with GTP-bound Rab5 and the Mon1-Ccz1 complex (but not either protein alone) binds Rab7 and also influences Rab7 activation (Kinchen and Ravichandran 2010). Yeast Mon1-Ccz1 complex was found to possess a novel GEF activity toward Ypt7, the yeast homolog of Rab7 (Nordmann et al. 2010). It is possible that in C. elegans, active RAB-5 on the phagosomal surfaces recruits the SAND-1-CCZ-1 complex by interacting with SAND-1, and the SAND-1-CCZ-1 complex, in turn, recruits and activates RAB-7 via its putative GEF activity to convert the phagosomes from the RAB-5-positive early to RAB-7-positive late stages.
Homotypic fusion and protein sorting complex—a potential RAB-7 effector in phagosome maturation:
The homotypic fusion and protein sorting (HOPS) complex; comprised of six subunits, VPS11, 16, 18, 33, 39, and 41; was previously identified in yeast as a complex that promotes homotypic vacuole fusion (Nickerson et al. 2009). Inactivation of any component of the HOPS complex in C. elegans; including vps-11, vps-16, vps-18, vps-33.1, vps-39, vps-41, or vps-45; results in failure of cell corpse degradation (Kinchen et al. 2008; Xiao et al. 2009). Characterization of the vps-18 mutant suggests that vps-18 is required for the biogenesis of endosomes and lysosomes and for the fusion of apoptotic cell-containing phagosomes with lysosomes (Xiao et al. 2009). Although the HOPS complex has been reported to be a GEF for the yeast Rab7 Ypt7; in C. elegans mutants defective in HOPS complex formation, apoptotic cell-containing phagosomes are arrested at the RAB-7-positive stage (Kinchen et al. 2008; Akbar et al. 2011). Therefore, the HOPS complex likely acts downstream of RAB-7 in C. elegans. The HOPS component VPS-41 interacts with the lysosome-localized small GTPase ARF-like 8 (ARL-8) (Sasaki et al. 2013). In arl-8 mutants, apoptotic cell-containing phagosomes fail to fuse with lysosomes and are arrested at the RAB-7-positive stage (Sasaki et al. 2013). Thus, HOPS may mediate the fusion of RAB-7-positive phagosomes to lysosomes through, at least in part, the interaction between VPS-41 and ARL-8.
Lipid second messenger PtdIns(3)P and its effector proteins:
After sealing, PtdIns(3)P is enriched on maturing phagosomes in an oscillating pattern with two bursts of PtdIns(3)P signals (Kinchen et al. 2008; Mangahas et al. 2008; Yu et al. 2008). This dynamic PtdIns(3)P pattern is achieved by sequential and combined action of PIKI-1 and VPS-34 (Lu et al. 2012). PIKI-1 is required for the initial production of PtdIns(3)P, whereas VPS-34 is needed for the generation of PtdIns(3)P in both the second half of the first wave and the second wave (Lu et al. 2012; Cheng et al. 2015). Lu et al. (2012) reported that MTM-1 antagonizes the activities of PIKI-1 and VPS-34 on phagosomes and results in the gap period between the two PtdIns(3)P waves on phagosomes. They proposed that timely dephosphorylation of PtdIns(3)P might be critical for the dissociation of certain initial phagosome maturation factors from phagosomes, as well as the subsequent association of maturation factors that act at later stages of phagosome maturation. However, Cheng et al.(2015) found that PtdIns(3)P waves associate with clearance of some, but not all, corpses, and do not involve functions of MTM-1.The role of PtdIns(3)P waves in phagosome maturation needs further investigation.
Like LST-4, sorting nexin 1 (SNX-1) and SNX-6, which contain the PtdIns(3)P-binding phox homology domain and belong to a family of sorting nexins, have been identified as PtdIns(3)P effectors during cell corpse clearance (N. Lu et al. 2011; D. Chen et al. 2013). SNX-1, SNX-6, and their human homologs are components of the retromer complex that mediates endosome-to-Golgi retrieval of transmembrane receptors and other trafficking-related proteins in worms and humans (Coudreuse et al. 2006; Verges 2007; Pan et al. 2008; Yang et al. 2008; Shi et al. 2009). The engulfment receptor CED-1 has been reported to be recycled from the phagosome back to the plasma membrane by the retromer complex. Loss of snx-1 or snx-6, but not lst-4, reduces CED-1 protein levels and inhibits recycling of CED-1 from phagosomes to the plasma membrane, resulting in lysosomal degradation of the receptor (Chen et al. 2010; Harterink et al. 2011). This result is further supported by the observation that perturbing phagosomal PtdIns(3)P levels led to CED-1 persistence on phagosomes (Neukomm et al. 2011b), given that SNX-1 and SNX-6 are PtdIns(3)P effectors. Thus, SNX-1 and SNX-6 may bind to PtdIns(3)P and act as PtdIns(3)P effectors to mediate CED-1 recycling (Figure 4B) (Chen et al. 2010; N. Lu et al. 2011). However, a different study found that the level of CED-1 on engulfing cell surfaces is not primarily controlled by SNX-1 or SNX-6 (N. Lu et al. 2011).
In addition, SNX-1, SNX-6, and LST-4 have been reported to mediate multiple events during phagosome maturation, including the fusion of endosomes and lysosomes to phagosomes. Genetic studies suggest that snx-1 and snx-6 function in the same pathway during phagosome maturation and that lst-4 acts in another pathway in a partially-redundant manner (N. Lu et al. 2011). SNX-1 physically interacts with SNX-6 and recruits SNX-6 to phagosomes (N. Lu et al. 2011). Consistent with the idea that LST-4 acts in a different pathway from SNX-1 and SNX-6, it is reported that LST-4, but not SNX-1 or SNX-6, is required for sealing of phagosomes (Cheng et al. 2015); and that SNX-1 and SNX-6, but not LST-4, are involved in CED-1 receptor recycling (D. Chen et al. 2013) and phagosomal accumulation of PtdIns(3)P, which recruits SNX-1 and SNX-6, following the dissociation of LST-4 (Almendinger et al. 2011; D. Chen et al. 2013; Cheng et al. 2015). However, N. Lu et al. (2011) found that SNX-1 and LST-4 function in a similar manner as they both associate with phagosomal surfaces and were particularly enriched on phagosomal tubules and the base of the tubules, despite that the identity of phagosomal tubules have not yet been determined. Mutation in snx-1 and/or lst-4 reduced the efficiency of the delivery of lysosomes into phagosomes; probably due to defects in the formation of phagosomal tubules, which function to capture and recruit cytosolic lysosomes to the surface of phagosomes (Harrison et al. 2003; Yu et al. 2008). In addition, the formation of phagosomal tubule depends on the Bin-Amphiphysin-Rvs domain of SNX-1 and LST-4 (N. Lu et al. 2011). Based on these results and the mutually-dependent localization of LST-4 and DYN-1 on phagosomal surface, it was proposed that SNX-1, SNX-6, and LST-4 promote phagosome maturation through regulating phagosomal tubule extension and stabilizing DYN-1 binding to phagosomal surfaces (Almendinger et al. 2011; N. Lu et al. 2011). It is intriguing that LST-4, which functions in scission of phagosomes as a fission factor (Cheng et al. 2015), also promotes fusion of vesicles along phagosomal tubules during phagosomal maturation (N. Lu et al. 2011).
Autophagy genes:
Recent analyses of autophagy genes revealed their functions in the degradation of apoptotic cells (Li et al. 2012; Cheng et al. 2013; S. Huang et al. 2013). Autophagy is a catabolic process through which protein aggregates and damaged organelles are delivered to lysosomes for degradation. lf mutations in several autophagy genes that act at distinct steps in the autophagy pathway caused accumulation of cell corpses and delayed cell corpse clearance (Cheng et al. 2013; S. Huang et al. 2013). Further analysis of some of these autophagy mutants suggested that they act in parallel to PIKI-1 to regulate phagosomal PtdIns3P levels in a similar manner as VPS-34 during phagosome maturation (Cheng et al. 2013). In addition, autophagy proteins ectopic P granule 5 (EPG-5); autophagy 18 (ATG-18) (yeast Atg homolog); and LC3, GABARAP and GATE-16 family 1 (LGG-1) are recruited to phagosomes in a stepwise order during degradation of germ cell corpses, but not embryonic cell corpses. This suggests that the three proteins function at distinct steps of phagosomal maturation during degradation of germ cell corpses, and that distinct mechanisms may be employed during maturation of corpse-containing phagosomes in embryos and adult germline (Cheng et al. 2013; S. Huang et al. 2013).
Acidification of phagosomal lumen
Phagosome maturation is accompanied by progressive acidification of phagosomal lumen, as lysosomal enzymes that degrade phagosomal contents prefer an acidic condition (Beyenbach and Wieczorek 2006; Steinberg et al. 2007). Acidification of phagosomes in C. elegans requires specific small GTPases, RAB-2 and RAB-14, which act in a partially-redundant manner, but not RAB-7 (Mangahas et al. 2008; Yu et al. 2008; Guo et al. 2010). The observation that mutations in rab-7 or sand-1 block the fusion of phagosomes to lysosomes but does not affect the acidification of phagosomes suggests that acidification of phagosomes occurs independently of lysosomes. Consistently, phagosomal lumen has turned acidic (pH ∼ 5) before phagosomes fuse with lysosomes (McNeil et al. 1983). However, how phagosomal lumen is acidified is not yet understood. Acidification of lysosomal lumen is important for apoptotic cell degradation. Inactivation of the vacuolar H ATPase 12 (vha-12) gene encoding the V1 B subunit of the lysosomal proton pump vacuolar-type ATPase (V-ATPase), impairs the degradation of apoptotic cells (Ernstrom et al. 2012).
Digestion of apoptotic cells
Once a cell undergoes the execution phase of apoptosis, several morphological changes occur, including cell shrinkage, membrane blebbing, chromosome condensation, and nuclear DNA fragmentation (Kerr et al. 1972; Wyllie 1980; Oberhammer et al. 1993). These processes can occur in apoptotic cells with little contribution from engulfing cells. Once internalized by engulfing cells, apoptotic cells are enclosed in phagosomes, which sequentially mature to phagolysosomes. The apoptotic cells are eventually digested by enzymes in phagolysosomes provided by lysosomes in engulfing cells.
Degradation of proteins of apoptotic cells:
The CED-3 caspase is the major protease that is activated to promote apoptosis and cause disassembly of the dying cell (see above). The apoptotic cells are further digested by proteases in phagolysosomes and cathepsin-like 1 (CPL-1) appears to be the major lysosomal protease for apoptotic cell degradation (Xu et al. 2014). Among 43 lysosomal proteases tested, inactivation of the cysteine protease CPL-1, but not others, significantly impairs degradation of apoptotic cells. CPL-1 does not affect the phagosomal maturation processes, including phagosomal recruitment of RAB-5, RAB-7, or LST-4, and phagosome acidification (Xu et al. 2014).
Degradation of DNA of apoptotic cells:
The degradation of chromosome DNA of apoptotic cells involves autonomous nucleases of the dying cell, nucleases secreted from other cells, and the lysosomal nuclease(s) of an engulfing cell. These nucleases act in sequential steps, in different stages, and in different sites to generate DNA intermediates of different sizes and to complete the degradation process (Parrish and Xue 2006). As discussed above, the activation of the CED-3 caspase results in the cleavage and activation of the DCR-1 nuclease that makes the first cuts on chromosomes (Nakagawa et al. 2010). The CED-3-mediated release of the mitochondrial protein WAH-1 and the mitochondrial endonuclease CPS-6, and their translocation to the nucleus, lead to the assembly of the multi-nuclease degradeosome, including multiple CRN nucleases, which appears to be the major workforce in fragmenting chromosomal DNA (Parrish et al. 2001; Wang et al. 2002; Parrish and Xue 2003). NUC-1, CRN-6, and CRN-7, three C. elegans DNase II endonucleases, catalyze the late stage DNA degradation (Wu et al. 2000; Parrish and Xue 2003), with NUC-1 constituting the major DNase II activity during apoptotic DNA degradation and CRN-6 and CRN-7 playing an auxiliary and a negligible role, respectively (Lai et al. 2009). Unlike DNase II in mammals, which is thought to function in engulfing cells, NUC-1 and CRN-6 may act as secreted endonucleases and function in apoptotic cells rather than in phagocytes for apoptotic DNA degradation. It is proposed that NUC-1 and CRN-6 are secreted from a distance and retaken up by apoptotic cells to promote DNA degradation.
It is interesting that apoptotic DNA degradation is required not only for the proper progression of apoptosis but it is also important for efficient cell corpse engulfment. Inactivation of both cps-6 and crn-2 increases the duration time of embryonic cell corpses (Parrish and Xue 2003). When released from mitochondria during apoptosis, WAH-1 promotes PS externalization and therefore cell corpse engulfment (Wang et al. 2007). Thus, the distinct steps of apoptosis including execution, engulfment, and DNA degradation are highly coordinated to ensure the efficient progression of cell death.
Nonapoptotic Cell Death
As mentioned in the Introduction, the majority of the 131 programmed cell deaths that occur during the development of the C. elegans soma depend on the core apoptotic machinery (EGL-1 BH3-only, CED-9 Bcl-1, CED-4 Apaf-1, CED-3 Caspase) (Horvitz 2003; Lettre and Hengartner 2006; Conradt 2009). However, the programmed death of the linker cell in males is independent of this machinery. Furthermore, the engulfment of the linker cell corpse is independent of known engulfment genes (Ellis and Horvitz 1986; Abraham et al. 2007; Blum et al. 2012). Consistent with the notion that the death of the linker cell occurs through a nonapoptotic process, dying linker cells do not display the morphological features that are characteristic of apoptotic cells that die during C. elegans development (Abraham et al. 2007; Blum et al. 2012). Dying linker cells maintain the morphology of healthy cells even during the engulfment process and no condensation of chromatin can be observed in the nucleus. At a later stage, the nucleus becomes increasingly indented or “crenellated.” In addition, cytoplasmic membrane structures can be observed that are not detected in healthy cells, and that may include swollen mitochondria within large multilayered membrane-bound structures. Some of these structures are reminiscent of structures observed in necrotic cells and/or autophagic vesicles; however, the linker cell death is also independent of genes that have been implicated in necrosis and autophagy (Abraham et al. 2007). Interestingly, similar morphological features have been observed in two different contexts in the vertebrate nervous system: in neurons that die during the development of the spinal cord and ciliary ganglia, and during polyglutamine-induced neurodegeneration. This suggests that this nonapoptotic form of programmed cell death is conserved from C. elegans to mammals (Abraham et al. 2007; Blum et al. 2012).
What is known about the molecular machineries that control and execute the programmed death of the linker cell? The linker cell is born during the second larval stage (L2 stage) and it dies at the transition from the last larval stage (L4 stage) to adulthood (Sulston et al. 1983). Unlike most cells that are programmed to die, the linker cell differentiates, migrates, and fulfills a function (to lead the extension of the developing male gonad) before it dies. Mutations in the heterochronic pathway that disrupt the L4/adult transition [such as lf mutations of the gene lin-29, which encodes a Zn-finger transcription factor, and lf mutations of the microRNA gene lethal 7 (let-7)] cause up to ∼50% of linker cells to inappropriately survive, indicating that correct developmental timing is critical for the specification of the linker cell death (Abraham et al. 2007). Furthermore, the gene prion-like-(Q/N-rich)-domain-bearing protein 41 (pqn-41), which encodes a polyglutamine repeat-containing protein, is required for the death of ∼20% of the linker cells (Blum et al. 2012). pqn-41 acts cell autonomously (i.e., in the linker cell) and is transcriptionally upregulated at the onset of linker cell death. For these reasons, pqn-41 may be a component of the molecular machinery that activates and/or executes linker cell death. Also required for linker cell death are the genes troll and interleukin 1 receptor domain protein 1 (tir-1) and SAPK/ERK kinase 1 (sek-1), which encode a p38 MAPK scaffolding protein and a p38 cascade MAPK kinase, respectively (Blum et al. 2012). tir-1 and sek-1 are known for their functions in C. elegans innate immunity, neuronal differentiation, and stress response; however, in the context of the linker cell death, tir-1 and sek-1 appear to act independently of these processes. Interestingly, sek-1 and lin-29 both act in the linker cell (however in two parallel pathways) to cause transcriptional upregulation of pqn-41 at the onset of linker cell death (Blum et al. 2012). This suggests that the death of the linker cell is specified by multiple signals, which may converge on a common nonapoptotic cell death machinery. One component of this cell death machinery may be the endogenous polyglutamine-repeat containing protein PQN-41.
Conclusions
The genetic, molecular, cell biological, and biochemical characterization of genes that affect developmental cell death in C. elegans has revealed numerous exciting molecular mechanisms involved in cell death activation, specification, and execution. There are still many questions regarding programmed cell death to be addressed. For example, while we know that the transcriptional activation of the egl-1 gene specifies whether a cell will live or die in many cells, we are far from having a comprehensive understanding of how egl-1 expression is controlled. Furthermore, while we have improved our understanding of how CED-3 is activated during apoptosis and what CED-3 substrates are, there are probably additional positive and negative regulators of the cell killing machinery and additional physiological CED-3 substrates that are important for other cell death execution events, such as cell corpse clearance, cytoplasm shrinkage, and nuclear membrane breakdown. How the release of mitochondrial apoptogenic factors, such as cytochrome c and AIF, is tightly regulated during apoptosis remains a major unsolved issue in the field of apoptosis. The powerful genetic and reverse genetic tools available in C. elegans should provide a unique genetic approach to unravel this long-standing question. Finally, while much more is known about the molecular components that act in the phagocytes to mediate cell corpse engulfment and degradation, little is known about what acts in the dying cell to trigger the phagocytosis event and how engulfing cells promote cell killing in a cell nonautonomous manner. By answering these and other remaining questions, studies of developmental cell death in C. elegans will continue to contribute in a major way to our current knowledge of programmed cell death.
Acknowledgments
We thank Yi-Ting Cheng for producing Figure 4. Research in D.X.’s laboratory is supported by grants from the National Institutes of Health (R01 GM-59083, R01 GM-79097, and R35 GM-118188). Research in B.C.’s laboratory is supported by funds from the Deutsche Forschungsgemeinschaft (CO 204/6-1, CRC 646, and EXC 114). Research in Y.-C.W.’s laboratory is supported by Ministry of Science and Technology grants (102-2311-B-002-047-MY3 and 104-2627-M-002-001) and a National Taiwan University grant (104R7602A3).
Footnotes
Communicating editor: M. V. Sundaram
Literature Cited
- Abraham M. C., Lu Y., Shaham S., 2007. A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev. Cell 12: 73–86. [DOI] [PubMed] [Google Scholar]
- Adams J. M., 2003. Ways of dying: multiple pathways to apoptosis. Genes Dev. 17: 2481–2495. [DOI] [PubMed] [Google Scholar]
- Adams J. M., Cory S., 2001. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem. Sci. 26: 61–66. [DOI] [PubMed] [Google Scholar]
- Akbar M. A., Tracy C., Kahr W. H., Kramer H., 2011. The full-of-bacteria gene is required for phagosome maturation during immune defense in Drosophila. J. Cell Biol. 192: 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendinger J., Doukoumetzidis K., Kinchen J. M., Kaech A., Ravichandran K. S., et al. , 2011. A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PLoS One 6: e18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnemri E. S., Livingston D. J., Nicholson D. W., Salvesen G., Thornberry N. A., et al. , 1996. Human ICE/CED-3 protease nomenclature. Cell 87: 171. [DOI] [PubMed] [Google Scholar]
- Anderson C., Zhou S., Sawin E., Horvitz H. R., Hurwitz M. E., 2012. SLI-1 Cbl inhibits the engulfment of apoptotic cells in C. elegans through a ligase-independent function. PLoS Genet. 8: e1003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends M. J., Morris R. G., Wyllie A. H., 1990. Apoptosis. The role of the endonuclease. Am. J. Pathol. 136: 593–608. [PMC free article] [PubMed] [Google Scholar]
- Arnoult D., Rismanchi N., Grodet A., Roberts R. G., Seeburg D. P., et al. , 2005. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr. Biol. 15: 2112–2118. [DOI] [PubMed] [Google Scholar]
- Avery L., Horvitz H. R., 1987. A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell 51: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A., Gartner A., 2013. Germ cell apoptosis and DNA damage responses. Adv. Exp. Med. Biol. 757: 249–276. [DOI] [PubMed] [Google Scholar]
- Baker M., Mackenzie I. R., Pickering-Brown S. M., Gass J., Rademakers R., et al. , 2006. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442: 916–919. [DOI] [PubMed] [Google Scholar]
- Bateman A., Bennett H. P., 2009. The granulin gene family: from cancer to dementia. BioEssays 31: 1245–1254. [DOI] [PubMed] [Google Scholar]
- Beyenbach K. W., Wieczorek H., 2006. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209: 577–589. [DOI] [PubMed] [Google Scholar]
- Bloss T. A., Witze E. S., Rothman J. H., 2003. Suppression of CED-3-independent apoptosis by mitochondrial betaNAC in Caenorhabditis elegans. Nature 424: 1066–1071. [DOI] [PubMed] [Google Scholar]
- Blum E. S., Abraham M. C., Yoshimura S., Lu Y., Shaham S., 2012. Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science 335: 970–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohdanowicz M., Grinstein S., 2013. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol. Rev. 93: 69–106. [DOI] [PubMed] [Google Scholar]
- Bohdanowicz M., Balkin D. M., De Camilli P., Grinstein S., 2012. Recruitment of OCRL and Inpp5B to phagosomes by Rab5 and APPL1 depletes phosphoinositides and attenuates Akt signaling. Mol. Biol. Cell 23: 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose J., Gruber A. D., Helming L., Schiebe S., Wegener I., et al. , 2004. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P., Strasser A., 2002. BH3-only proteins - evolutionarily conserved proapoptotic Bcl-2 family members essential for initiating programmed cell death. J. Cell Sci. 115: 1567–1574. [DOI] [PubMed] [Google Scholar]
- Breckenridge D. G., Kang B. H., Kokel D., Mitani S., Staehelin L. A., et al. , 2008. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol. Cell 31: 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 2003. Nobel lecture. Nature’s gift to science. Biosci. Rep. 23: 225–237. [DOI] [PubMed] [Google Scholar]
- Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., et al. , 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4: 574–582. [DOI] [PubMed] [Google Scholar]
- Budihardjo I., Oliver H., Lutter M., Luo X., Wang X., 1999. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15: 269–290. [DOI] [PubMed] [Google Scholar]
- Cabello J., Neukomm L. J., Gunesdogan U., Burkart K., Charette S. J., et al. , 2010. The Wnt pathway controls cell death engulfment, spindle orientation, and migration through CED-10/Rac. PLoS Biol. 8: e1000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello J., Samann J., Gomez-Orte E., Erazo T., Coppa A., et al. , 2014. PDR-1/hParkin negatively regulates the phagocytosis of apoptotic cell corpses in Caenorhabditis elegans. Cell Death Dis. 5: e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S., Clark S. G., McDermott J. B., Aamodt E., Horvitz H. R., 2002. PAG-3, a Zn-finger transcription factor, determines neuroblast fate in C. elegans. Development 129: 1763–1774. [DOI] [PubMed] [Google Scholar]
- Cereghetti G. M., Scorrano L., 2006. The many shapes of mitochondrial death. Oncogene 25: 4717–4724. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Lambie E. J., Bindu S., Mikeladze-Dvali T., Conradt B., 2015. Engulfment pathways promote programmed cell death by enhancing the unequal segregation of apoptotic potential. Nat. Commun. 6: 10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Horvitz H. R., Sulston J. E., 1981. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24: 59–69. [DOI] [PubMed] [Google Scholar]
- Chen D., Xiao H., Zhang K., Wang B., Gao Z., et al. , 2010. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science 327: 1261–1264. [DOI] [PubMed] [Google Scholar]
- Chen D., Jian Y., Liu X., Zhang Y., Liang J., et al. , 2013. Clathrin and AP2 are required for phagocytic receptor-mediated apoptotic cell clearance in Caenorhabditis elegans. PLoS Genet. 9: e1003517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Hersh B. M., Conradt B., Zhou Z., Riemer D., et al. , 2000. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 287: 1485–1489. [DOI] [PubMed] [Google Scholar]
- Chen Y. Z., Mapes J., Lee E. S., Skeen-Gaar R. R., Xue D., 2013. Caspase-mediated activation of Caenorhabditis elegans CED-8 promotes apoptosis and phosphatidylserine externalization. Nat. Commun. 4: 2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Wu Y., Lu Q., Yan J., Zhang H., et al. , 2013. Autophagy genes coordinate with the class II PI/PtdIns 3-kinase PIKI-1 to regulate apoptotic cell clearance in C. elegans. Autophagy 9: 2022–2032. [DOI] [PubMed] [Google Scholar]
- Cheng S., Wang K., Zou W., Miao R., Huang Y., et al. , 2015. PtdIns(4,5)P(2) and PtdIns3P coordinate to regulate phagosomal sealing for apoptotic cell clearance. J. Cell Biol. 210: 485–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien S. C., Brinkmann E. M., Teuliere J., Garriga G., 2013. Caenorhabditis elegans PIG-1/MELK Acts in a Conserved PAR-4/LKB1 Polarity Pathway to Promote Asymmetric Neuroblast Divisions. Genetics 193: 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan A. M., O’Rourke K., Lane B. R., Dixit V. M., 1997. Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death. Science 275: 1122–1126. [DOI] [PubMed] [Google Scholar]
- Chotard L., Mishra A. K., Sylvain M. A., Tuck S., Lambright D. G., et al. , 2010. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell 21: 2285–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. G., Shurland D. L., Meyerowitz E. M., Bargmann C. I., van der Bliek A. M., 1997. A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans. Proc. Natl. Acad. Sci. USA 94: 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G. M., 1997. Caspases: the executioners of apoptosis. Biochem. J. 326: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B., 2009. Genetic control of programmed cell death during animal development. Annu. Rev. Genet. 43: 493–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B., Horvitz H. R., 1998. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93: 519–529. [DOI] [PubMed] [Google Scholar]
- Conradt B., Horvitz H. R., 1999. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98: 317–327. [DOI] [PubMed] [Google Scholar]
- Cordes S., Frank C. A., Garriga G., 2006. The C. elegans MELK ortholog PIG-1 regulates cell size asymmetry and daughter cell fate in asymmetric neuroblast divisions. Development 133: 2747–2756. [DOI] [PubMed] [Google Scholar]
- Coudreuse D. Y., Roel G., Betist M. C., Destree O., Korswagen H. C., 2006. Wnt gradient formation requires retromer function in Wnt-producing cells. Science 312: 921–924. [DOI] [PubMed] [Google Scholar]
- Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., et al. , 2006. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442: 920–924. [DOI] [PubMed] [Google Scholar]
- Cully M., You H., Levine A. J., Mak T. W., 2006. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer 6: 184–192. [DOI] [PubMed] [Google Scholar]
- Danial N. N., Korsmeyer S. J., 2004. Cell death: critical control points. Cell 116: 205–219. [DOI] [PubMed] [Google Scholar]
- Darland-Ransom M., Wang X., Sun C. L., Mapes J., Gengyo-Ando K., et al. , 2008. Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science 320: 528–531. [DOI] [PubMed] [Google Scholar]
- deBakker C. D., Haney L. B., Kinchen J. M., Grimsley C., Lu M., et al. , 2004. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr. Biol. 14: 2208–2216. [DOI] [PubMed] [Google Scholar]
- del Peso L., Gonzalez V. M., Nunez G., 1998. Caenorhabditis elegans EGL-1 disrupts the interaction of CED-9 with CED-4 and promotes CED-3 activation. J. Biol. Chem. 273: 33495–33500. [DOI] [PubMed] [Google Scholar]
- Denning D. P., Hatch V., Horvitz H. R., 2012. Programmed elimination of cells by caspase-independent cell extrusion in C. elegans. Nature 488: 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. P., Hatch V., Horvitz H. R., 2013. Both the caspase CSP-1 and a caspase-independent pathway promote programmed cell death in parallel to the canonical pathway for apoptosis in Caenorhabditis elegans. PLoS Genet. 9: e1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P., 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657. [DOI] [PubMed] [Google Scholar]
- Ellis H. M., Horvitz H. R., 1986. Genetic control of programmed cell death in the nematode C. elegans. Cell 44: 817–829. [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Horvitz H. R., 1991. Two C. elegans genes control the programmed deaths of specific cells in the pharynx. Development 112: 591–603. [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Jacobson D. M., Horvitz H. R., 1991. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129: 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernstrom G. G., Weimer R., Pawar D. R., Watanabe S., Hobson R. J., et al. , 2012. V-ATPase V1 sector is required for corpse clearance and neurotransmission in Caenorhabditis elegans. Genetics 191: 461–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeel B., Xue D., 2009. The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit. Rev. Biochem. Mol. Biol. 44: 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok V. A., Bratton D. L., Frasch S. C., Warner M. L., Henson P. M., 1998. The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death Differ. 5: 551–562. [DOI] [PubMed] [Google Scholar]
- Fadok V. A., Bratton D. L., Rose D. M., Pearson A., Ezekewitz R. A., et al. , 2000. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 405: 85–90. [DOI] [PubMed] [Google Scholar]
- Fadok V. A., Savill J. S., Haslett C., Bratton D. L., Doherty D. E., et al. , 1992a Different populations of macrophages use either the vitronectin receptor or the phosphatidylserine receptor to recognize and remove apoptotic cells. J. Immunol. 149: 4029–4035. [PubMed] [Google Scholar]
- Fadok V. A., Voelker D. R., Campbell P. A., Cohen J. J., Bratton D. L., et al. , 1992b Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148: 2207–2216. [PubMed] [Google Scholar]
- Fadok V. A., Xue D., Henson P., 2001. If phosphatidylserine is the death knell, a new phosphatidylserine-specific receptor is the bellringer. Cell Death Differ. 8: 582–587. [DOI] [PubMed] [Google Scholar]
- Fancsalszky L., Monostori E., Farkas Z., Pourkarimi E., Masoudi N., et al. , 2014. NDK-1, the homolog of NM23–H1/H2 regulates cell migration and apoptotic engulfment in C. elegans. PLoS One 9: e92687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Horvitz H. R., 1988. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell 55: 757–769. [DOI] [PubMed] [Google Scholar]
- Frank C. A., Hawkins N. C., Guenther C., Horvitz H. R., Garriga G., 2005. C. elegans HAM-1 positions the cleavage plane and regulates apoptosis in asymmetric neuroblast divisions. Dev. Biol. 284: 301–310. [DOI] [PubMed] [Google Scholar]
- Fuchs Y., Steller H., 2011. Programmed cell death in animal development and disease. Cell 147: 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin B. D., Denning D. P., Horvitz H. R., 2011. SPK-1, an SR protein kinase, inhibits programmed cell death in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108: 1998–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner, A., P. R. Boag, and T. K. Blackwell, 2008 Germline survival and apoptosis (September 4, 2008), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.145.1, http://www.wormbook.org. 10.1895/wormbook.1.145.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O., 2000. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y., Sherman Y., Ben-Sasson S. A., 1992. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119: 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Zhao X., Nakagawa A., Gong X., Skeen-Gaar R. R., et al. , 2014. A novel mechanism underlies caspase-dependent conversion of the dicer ribonuclease into a deoxyribonuclease during apoptosis. Cell Res. 24: 218–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X., Shi Y., Nakagawa A., Yoshina S., Mitani S., et al. , 2008. Inhibition of CED-3 zymogen activation and apoptosis in Caenorhabditis elegans by caspase homolog CSP-3. Nat. Struct. Mol. Biol. 15: 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X., Zhou Q. H., Kage-Nakadai E., Shi Y., Yan N., et al. , 2009. Caenorhabditis elegans caspase homolog CSP-2 inhibits CED-3 autoactivation and apoptosis in germ cells. Cell Death Differ. 16: 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther C., Garriga G., 1996. Asymmetric distribution of the C. elegans HAM-1 protein in neuroblasts enables daughter cells to adopt distinct fates. Development 122: 3509–3518. [DOI] [PubMed] [Google Scholar]
- Gumienny T. L., Lambie E., Hartwieg E., Horvitz H. R., Hengartner M. O., 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Gumienny T. L., Brugnera E., Tosello-Trampont A. C., Kinchen J. M., Haney L. B., et al. , 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107: 27–41. [DOI] [PubMed] [Google Scholar]
- Guo P., Hu T., Zhang J., Jiang S., Wang X., 2010. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc. Natl. Acad. Sci. USA 107: 18016–18021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurling M., Talavera K., Garriga G., 2014. The DEP domain-containing protein TOE-2 promotes apoptosis in the Q lineage of C. elegans through two distinct mechanisms. Development 141: 2724–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon Y., Trompier D., Ma Z., Venegas V., Pophillat M., et al. , 2006. Cooperation between engulfment receptors: the case of ABCA1 and MEGF10. PLoS One 1: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation vs. meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 71–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. E., Bucci C., Vieira O. V., Schroer T. A., Grinstein S., 2003. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol. Cell. Biol. 23: 6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M., Port F., Lorenowicz M. J., McGough I. J., Silhankova M., et al. , 2011. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13: 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzold J., Conradt B., 2008. Control of apoptosis by asymmetric cell division. PLoS Biol. 6: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Lu N., Zhou Z., 2009. Cellular and nuclear degradation during apoptosis. Curr. Opin. Cell Biol. 21: 900–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Yu X., Margolis M., Liu X., Leng X., et al. , 2010. Live-cell imaging in Caenorhabditis elegans reveals the distinct roles of dynamin self-assembly and guanosine triphosphate hydrolysis in the removal of apoptotic cells. Mol. Biol. Cell 21: 610–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., Sulston J. E., Thomson J. N., 1983. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220: 1277–1279. [DOI] [PubMed] [Google Scholar]
- Hengartner M. O., Horvitz H. R., 1994a Activation of C. elegans cell death protein CED-9 by an amino-acid substitution in a domain conserved in Bcl-2. Nature 369: 318–320. [DOI] [PubMed] [Google Scholar]
- Hengartner M. O., Horvitz H. R., 1994b C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 76: 665–676. [DOI] [PubMed] [Google Scholar]
- Hengartner M. O., Ellis R. E., Horvitz H. R., 1992. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356: 494–499. [DOI] [PubMed] [Google Scholar]
- Hirose T., Horvitz H. R., 2013. An Sp1 transcription factor coordinates caspase-dependent and -independent apoptotic pathways. Nature 500: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Horvitz H. R., 2014. The translational regulators GCN-1 and ABCF-3 act together to promote apoptosis in C. elegans. PLoS Genet. 10: e1004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Galvin B. D., Horvitz H. R., 2010. Six and Eya promote apoptosis through direct transcriptional activation of the proapoptotic BH3-only gene egl-1 in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107: 15479–15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatomi T., Sakamoto T., Sonoda K. H., Tsutsumi C., Qiao H., et al. , 2003. Clearance of apoptotic photoreceptors: elimination of apoptotic debris into the subretinal space and macrophage-mediated phagocytosis via phosphatidylserine receptor and integrin alphavbeta3. Am. J. Pathol. 162: 1869–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochreiter-Hufford A., Ravichandran K. S., 2013. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb. Perspect. Biol. 5: a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., 1987. A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1: 731–745. [DOI] [PubMed] [Google Scholar]
- Hoeppner D. J., Hengartner M. O., Schnabel R., 2001. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412: 202–206. [DOI] [PubMed] [Google Scholar]
- Hong J. R., Lin G. H., Lin C. J., Wang W. P., Lee C. C., et al. , 2004. Phosphatidylserine receptor is required for the engulfment of dead apoptotic cells and for normal embryonic development in zebrafish. Development 131: 5417–5427. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., 1999. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 59: 1701s–1706s. [PubMed] [Google Scholar]
- Horvitz H. R., 2003. Worms, life, and death (Nobel lecture). ChemBioChem 4: 697–711. [DOI] [PubMed] [Google Scholar]
- Hsieh H. H., Hsu T. Y., Jiang H. S., Wu Y. C., 2012. Integrin alpha PAT-2/CDC-42 signaling is required for muscle-mediated clearance of apoptotic cells in Caenorhabditis elegans. PLoS Genet. 8: e1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. Y., Wu Y. C., 2010. Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr. Biol. 20: 477–486. [DOI] [PubMed] [Google Scholar]
- Huang C. Y., Chen J. Y., Wu S. C., Tan C. H., Tzeng R. Y., et al. , 2012. C. elegans EIF-3.K promotes programmed cell death through CED-3 caspase. PLoS One 7: e36584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Jia K., Wang Y., Zhou Z., Levine B., 2013. Autophagy genes function in apoptotic cell corpse clearance during C. elegans embryonic development. Autophagy 9: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Jiang T., Choi W., Qi S., Pang Y., et al. , 2013. Mechanistic insights into CED-4-mediated activation of CED-3. Genes Dev. 27: 2039–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz M. E., Vanderzalm P. J., Bloom L., Goldman J., Garriga G., et al. , 2009. Abl kinase inhibits the engulfment of apoptotic [corrected] cells in Caenorhabditis elegans. PLoS Biol. 7: e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Inukai T., Yoshihara T., Seyschab H., Ashmun R. A., et al. , 1996. Reversal of apoptosis by the leukaemia-associated E2A-HLF chimaeric transcription factor. Nature 382: 541–544. [DOI] [PubMed] [Google Scholar]
- Inoue A., Seidel M. G., Wu W., Kamizono S., Ferrando A. A., et al. , 2002. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cell 2: 279–288. [DOI] [PubMed] [Google Scholar]
- Inukai T., Inoue A., Kurosawa H., Goi K., Shinjyo T., et al. , 1999. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol. Cell 4: 343–352. [DOI] [PubMed] [Google Scholar]
- Jagasia R., Grote P., Westermann B., Conradt B., 2005. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature 433: 754–760. [DOI] [PubMed] [Google Scholar]
- Jiang H. S., Wu Y. C., 2014. LIN-3/EGF promotes the programmed cell death of specific cells in Caenorhabditis elegans by transcriptional activation of the pro-apoptotic gene egl-1. PLoS Genet. 10: e1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Wang X., 2004. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 73: 87–106. [DOI] [PubMed] [Google Scholar]
- Juhasz G., Hill J. H., Yan Y., Sass M., Baehrecke E. H., et al. , 2008. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 181: 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Xu J., Liu Y., Sun J., Sun D., et al. , 2011. Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 410: 189–194. [DOI] [PubMed] [Google Scholar]
- Kao A. W., Eisenhut R. J., Martens L. H., Nakamura A., Huang A., et al. , 2011. A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proc. Natl. Acad. Sci. USA 108: 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. F., Wyllie A. H., Currie A. R., 1972. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26: 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J. M., Ravichandran K. S., 2010. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 464: 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J. M., Cabello J., Klingele D., Wong K., Feichtinger R., et al. , 2005. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 434: 93–99. [DOI] [PubMed] [Google Scholar]
- Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., et al. , 2008. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinet M. J., Shaham S., 2014. Noncanonical cell death in the nematode Caenorhabditis elegans. Methods Enzymol. 545: 157–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., et al. , 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608. [DOI] [PubMed] [Google Scholar]
- Kitano M., Nakaya M., Nakamura T., Nagata S., Matsuda M., 2008. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature 453: 241–245. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y., Masuko S., Noda M., Inayoshi A., Sanui T., et al. , 2004. Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the phosphatidylserine receptor. Blood 103: 3362–3364. [DOI] [PubMed] [Google Scholar]
- Lai H. J., Lo S. J., Kage-Nakadai E., Mitani S., Xue D., 2009. The roles and acting mechanism of Caenorhabditis elegans DNase II genes in apoptotic dna degradation and development. PLoS One 4: e7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G., Hengartner M. O., 2006. Developmental apoptosis in C. elegans: a complex CEDnario. Nat. Rev. Mol. Cell Biol. 7: 97–108. [DOI] [PubMed] [Google Scholar]
- Li L. Y., Luo X., Wang X., 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412: 95–99. [DOI] [PubMed] [Google Scholar]
- Li M. O., Sarkisian M. R., Mehal W. Z., Rakic P., Flavell R. A., 2003. Phosphatidylserine receptor is required for clearance of apoptotic cells. Science 302: 1560–1563. [DOI] [PubMed] [Google Scholar]
- Li W., Zou W., Zhao D., Yan J., Zhu Z., et al. , 2009. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development 136: 2445–2455. [DOI] [PubMed] [Google Scholar]
- Li W., Zou W., Yang Y., Chai Y., Chen B., et al. , 2012. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J. Cell Biol. 197: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Venegas V., Nagaoka Y., Morino E., Raghavan P., et al. , 2015. Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms. PLoS Genet. 11: e1005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Strauss T. J., Potts M. B., Cameron S., 2006. Direct regulation of egl-1 and of programmed cell death by the Hox protein MAB-5 and by CEH-20, a C. elegans homolog of Pbx1. Development 133: 641–650. [DOI] [PubMed] [Google Scholar]
- Liu Q. A., Hengartner M. O., 1998. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell 93: 961–972. [DOI] [PubMed] [Google Scholar]
- Liu Q. A., Hengartner M. O., 1999. Human CED-6 encodes a functional homologue of the Caenorhabditis elegans engulfment protein CED-6. Curr. Biol. 9: 1347–1350. [DOI] [PubMed] [Google Scholar]
- Liu X., Kim C. N., Yang J., Jemmerson R., Wang X., 1996. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86: 147–157. [DOI] [PubMed] [Google Scholar]
- Lu M., Kinchen J. M., Rossman K. L., Grimsley C., deBakker C., et al. , 2004. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat. Struct. Mol. Biol. 11: 756–762. [DOI] [PubMed] [Google Scholar]
- Lu M., Kinchen J. M., Rossman K. L., Grimsley C., Hall M., et al. , 2005. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 15: 371–377. [DOI] [PubMed] [Google Scholar]
- Lu N., Zhou Z., 2012. Membrane trafficking and phagosome maturation during the clearance of apoptotic cells. Int. Rev. Cell Mol. Biol. 293: 269–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N., Shen Q., Mahoney T. R., Liu X., Zhou Z., 2011. Three sorting nexins drive the degradation of apoptotic cells in response to PtdIns(3)P signaling. Mol. Biol. Cell 22: 354–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N., Shen Q., Mahoney T. R., Neukomm L. J., Wang Y., et al. , 2012. Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol. 10: e1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Zhang Y., Hu T., Guo P., Li W., et al. , 2008. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development 135: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Lu Y., Rolland S. G., Conradt B., 2011. A molecular switch that governs mitochondrial fusion and fission mediated by the BCL2-like protein CED-9 of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108: E813–E822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R., Carlsson S. R., 2009. SNX9 - a prelude to vesicle release. J. Cell Sci. 122: 5–11. [DOI] [PubMed] [Google Scholar]
- Luo J., Manning B. D., Cantley L. C., 2003. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 4: 257–262. [DOI] [PubMed] [Google Scholar]
- Mangahas P. M., Yu X., Miller K. G., Zhou Z., 2008. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J. Cell Biol. 180: 357–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapes J., Chen Y. Z., Kim A., Mitani S., Kang B. H., et al. , 2012. CED-1, CED-7, and TTR-52 regulate surface phosphatidylserine expression on apoptotic and phagocytic cells. Curr. Biol. 22: 1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. J., Reutelingsperger C. P., McGahon A. J., Rader J. A., van Schie R. C., et al. , 1995. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 182: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer C. W., Chiorazzi M., Shaham S., 2007. Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development 134: 1357–1368. [DOI] [PubMed] [Google Scholar]
- McNeil P. L., Tanasugarn L., Meigs J. B., Taylor D. L., 1983. Acidification of phagosomes is initiated before lysosomal enzyme activity is detected. J. Cell Biol. 97: 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzstein M. M., Horvitz H. R., 1999. The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol. Cell 4: 309–319. [DOI] [PubMed] [Google Scholar]
- Metzstein M. M., Hengartner M. O., Tsung N., Ellis R. E., Horvitz H. R., 1996. Transcriptional regulator of programmed cell death encoded by Caenorhabditis elegans gene ces-2. Nature 382: 545–547. [DOI] [PubMed] [Google Scholar]
- Mitchell J. E., Cvetanovic M., Tibrewal N., Patel V., Colamonici O. R., et al. , 2006. The presumptive phosphatidylserine receptor is dispensable for innate anti-inflammatory recognition and clearance of apoptotic cells. J. Biol. Chem. 281: 5718–5725. [DOI] [PubMed] [Google Scholar]
- Nakagawa A., Shi Y., Kage-Nakadai E., Mitani S., Xue D., 2010. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science 328: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A., Sullivan K. D., Xue D., 2014. Caspase-activated phosphoinositide binding by CNT-1 promotes apoptosis by inhibiting the AKT pathway. Nat. Struct. Mol. Biol. 21: 1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima T., Sekiguchi T., Kuraoka A., Fukushima K., Shibata Y., et al. , 1993. Molecular cloning of a human cDNA encoding a novel protein, DAD1, whose defect causes apoptotic cell death in hamster BHK21 cells. Mol. Cell. Biol. 13: 6367–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawa M., Kage-Nakadai E., Aiso S., Okamoto K., Mitani S., et al. , 2012. Reduced expression of BTBD10, an Akt activator, leads to motor neuron death. Cell Death Differ. 19: 1398–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme R., Grote P., Tomasi T., Loser S., Holzkamp H., et al. , 2010. Transcriptional upregulation of both egl-1 BH3-only and ced-3 caspase is required for the death of the male-specific CEM neurons. Cell Death Differ. 17: 1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm L. J., Frei A. P., Cabello J., Kinchen J. M., Zaidel-Bar R., et al. , 2011a Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nat. Cell Biol. 13: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm L. J., Nicot A. S., Kinchen J. M., Almendinger J., Pinto S. M., et al. , 2011b The phosphoinositide phosphatase MTM-1 regulates apoptotic cell corpse clearance through CED-5-CED-12 in C. elegans. Development 138: 2003–2014. [DOI] [PubMed] [Google Scholar]
- Neukomm L. J., Zeng S., Frei A. P., Huegli P. A., Hengartner M. O., 2014. Small GTPase CDC-42 promotes apoptotic cell corpse clearance in response to PAT-2 and CED-1 in C. elegans. Cell Death Differ. 21: 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B., Coakley S., Giordano-Santini R., Linton C., Lee E. S., et al. , 2015. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature 517: 219–222. [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., Brett C. L., Merz A. J., 2009. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr. Opin. Cell Biol. 21: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C., Almendinger J., Gysi S., Gomez-Orte E., Kaech A., et al. , 2010. ccz-1 mediates the digestion of apoptotic corpses in C. elegans. J. Cell Sci. 123: 2001–2007. [DOI] [PubMed] [Google Scholar]
- Nordmann M., Cabrera M., Perz A., Brocker C., Ostrowicz C., et al. , 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20: 1654–1659. [DOI] [PubMed] [Google Scholar]
- Oberhammer F., Wilson J. W., Dive C., Morris I. D., Hickman J. A., et al. , 1993. Apoptotic death in epithelial cells: cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 12: 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Stuurman N., D’Ambrosio M., Vale R. D., 2010. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science 330: 677–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C. L., Baum P. D., Gu M., Jorgensen E. M., Clark S. G., et al. , 2008. C. elegans AP-2 and retromer control Wnt signaling by regulating mig-14/Wntless. Dev. Cell 14: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parone P. A., Martinou J. C., 2006. Mitochondrial fission and apoptosis: an ongoing trial. Biochim. Biophys. Acta 1763: 522–530. [DOI] [PubMed] [Google Scholar]
- Parrish J., Metters H., Chen L., Xue D., 2000. Demonstration of the in vivo interaction of key cell death regulators by structure-based design of second-site suppressors. Proc. Natl. Acad. Sci. USA 97: 11916–11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish J., Li L., Klotz K., Ledwich D., Wang X., et al. , 2001. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412: 90–94. [DOI] [PubMed] [Google Scholar]
- Parrish J. Z., Xue D., 2003. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol. Cell 11: 987–996. [DOI] [PubMed] [Google Scholar]
- Parrish J. Z., Xue D., 2006. Cuts can kill: the roles of apoptotic nucleases in cell death and animal development. Chromosoma 115: 89–97. [DOI] [PubMed] [Google Scholar]
- Parrish J. Z., Yang C., Shen B., Xue D., 2003. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J. 22: 3451–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden E., Kimberly E., Gengyo-Ando K., Mitani S., Xue D., 2007. Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes Dev. 21: 3195–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts M. B., Cameron S., 2011. Cell lineage and cell death: Caenorhabditis elegans and cancer research. Nat. Rev. Cancer 11: 50–58. [DOI] [PubMed] [Google Scholar]
- Potts, M. B., D. P. Wang and S. Cameron, 2009 Trithorax, Hox, and TALE-class homeodomain proteins ensure cell survival through repression of the BH3-only gene egl-1. Dev Biol. 329: 374–385. [DOI] [PubMed]
- Pourkarimi E., Greiss S., Gartner A., 2012. Evidence that CED-9/Bcl2 and CED-4/Apaf-1 localization is not consistent with the current model for C. elegans apoptosis induction. Cell Death Differ. 19: 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Pang Y., Hu Q., Liu Q., Li H., et al. , 2010. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141: 446–457. [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Horvitz H. R., 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2: 131–136. [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Horvitz H. R., 2004. The Engulfment Process of Programmed Cell Death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 20: 193–221. [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Cameron S., Horvitz H. R., 2001. Phagocytosis promotes programmed cell death in C. elegans. Nature 412: 198–202. [DOI] [PubMed] [Google Scholar]
- Reed J. C., 1997. Double identity for proteins of the Bcl-2 family. Nature 387: 773–776. [DOI] [PubMed] [Google Scholar]
- Riedl S. J., Shi Y., 2004. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 5: 897–907. [DOI] [PubMed] [Google Scholar]
- Robertson A. M. G., Thomson J. N., 1982. Morphology of programmed cell death in the ventral nerve cord of Caenorhabditis elegans larvae. Development 67: 89–100. [Google Scholar]
- Sasaki A., Nakae I., Nagasawa M., Hashimoto K., Abe F., et al. , 2013. Arl8/ARL-8 functions in apoptotic cell removal by mediating phagolysosome formation in Caenorhabditis elegans. Mol. Biol. Cell 24: 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savill J., Fadok V., 2000. Corpse clearance defines the meaning of cell death. Nature 407: 784–788. [DOI] [PubMed] [Google Scholar]
- Savill J., Fadok V., Henson P., Haslett C., 1993. Phagocyte recognition of cells undergoing apoptosis. Immunol. Today 14: 131–136. [DOI] [PubMed] [Google Scholar]
- Schwartz H. T., Horvitz H. R., 2007. The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes Dev. 21: 3181–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K., Kurata S., Yanagihashi Y., Brummelkamp T. R., Matsuda F., et al. , 2014. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344: 1164–1168. [DOI] [PubMed] [Google Scholar]
- Seshagiri S., Miller L. K., 1997. Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis. Curr. Biol. 7: 455–460. [DOI] [PubMed] [Google Scholar]
- Shaham S., 1998. Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. J. Biol. Chem. 273: 35109–35117. [DOI] [PubMed] [Google Scholar]
- Shaham S., Horvitz H. R., 1996a An alternatively spliced C. elegans ced-4 RNA encodes a novel cell death inhibitor. Cell 86: 201–208. [DOI] [PubMed] [Google Scholar]
- Shaham S., Horvitz H. R., 1996b Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev. 10: 578–591. [DOI] [PubMed] [Google Scholar]
- Shaham S., Reddien P. W., Davies B., Horvitz H. R., 1999. Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics 153: 1655–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Qin F., Gao Z., Cui J., Xiao H., et al. , 2009. Adenine nucleotide translocator cooperates with core cell death machinery to promote apoptosis in Caenorhabditis elegans. Mol. Cell. Biol. 29: 3881–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., He B., Lu N., Conradt B., Grant B. D., et al. , 2013. Phagocytic receptor signaling regulates clathrin and epsin-mediated cytoskeletal remodeling during apoptotic cell engulfment in C. elegans. Development 140: 3230–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A., Sun L., Banerjee R., Tobin M., Zhang Y., et al. , 2009. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 28: 3290–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi A., Teuliere J., Talavera K., Cordes S., Ou G., et al. , 2011. The Arf GAP CNT-2 regulates the apoptotic fate in C. elegans asymmetric neuroblast divisions. Curr. Biol. 21: 948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulachev V. P., Bakeeva L. E., Chernyak B. V., Domnina L. V., Minin A. A., et al. , 2004. Thread-grain transition of mitochondrial reticulum as a step of mitoptosis and apoptosis. Mol. Cell. Biochem. 256–257: 341–358. [DOI] [PubMed] [Google Scholar]
- Spector M. S., Desnoyers S., Hoeppner D. J., Hengartner M. O., 1997. Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature 385: 653–656. [DOI] [PubMed] [Google Scholar]
- Stanfield G. M., Horvitz H. R., 2000. The ced-8 gene controls the timing of programmed cell deaths in C. elegans. Mol. Cell 5: 423–433. [DOI] [PubMed] [Google Scholar]
- Steinberg B. E., Huynh K. K., Grinstein S., 2007. Phagosomal acidification: measurement, manipulation and functional consequences. Biochem. Soc. Trans. 35: 1083–1087. [DOI] [PubMed] [Google Scholar]
- Steller H., 1995. Mechanisms and genes of cellular suicide. Science 267: 1445–1449. [DOI] [PubMed] [Google Scholar]
- Su H. P., Nakada-Tsukui K., Tosello-Trampont A. C., Li Y., Bu G., et al. , 2002. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J. Biol. Chem. 277: 11772–11779. [DOI] [PubMed] [Google Scholar]
- Sugimoto A., Hozak R. R., Nakashima T., Nishimoto T., Rothman J. H., 1995. dad-1, an endogenous programmed cell death suppressor in Caenorhabditis elegans and vertebrates. EMBO J. 14: 4434–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., 1976. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275: 287–297. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., 1988 Cell Lineage, pp. 123–155 in The nematode Caenorhabditis elegans, Vol. 17, edited by W. B. Wood. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sulston J. E., 2003. Caenorhabditis elegans: the cell lineage and beyond (Nobel lecture). ChemBioChem 4: 688–696. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1981. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 82: 41–55. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., White J. G., 1980. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol. 78: 577–597. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Susin S. A., Lorenzo H. K., Zamzami N., Marzo I., Snow B. E., et al. , 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397: 441–446. [DOI] [PubMed] [Google Scholar]
- Suzuki J., Denning D. P., Imanishi E., Horvitz H. R., Nagata S., 2013. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341: 403–406. [DOI] [PubMed] [Google Scholar]
- Teuliere J., Cordes S., Singhvi A., Talavera K., Garriga G., 2014. Asymmetric neuroblast divisions producing apoptotic cells require the cytohesin GRP-1 in Caenorhabditis elegans. Genetics 198: 229–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellmann M., Hatzold J., Conradt B., 2003. The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development 130: 4057–4071. [DOI] [PubMed] [Google Scholar]
- Venegas V., Zhou Z., 2007. Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol. Biol. Cell 18: 3180–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verges M., 2007. Retromer and sorting nexins in development. Front. Biosci. 12: 3825–3851. [DOI] [PubMed] [Google Scholar]
- Vieira O. V., Botelho R. J., Grinstein S., 2002. Phagosome maturation: aging gracefully. Biochem. J. 366: 689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos M., Tavernarakis N., 2010. Non-apoptotic cell death in Caenorhabditis elegans. Dev. Dyn. 239: 1337–1351. [DOI] [PubMed] [Google Scholar]
- Wang J., Chitturi J., Ge Q., Laskova V., Wang W., et al. , 2015. The C. elegans COE transcription factor UNC-3 activates lineage-specific apoptosis and affects neurite growth in the RID lineage. Development 142: 1447–1457. [DOI] [PubMed] [Google Scholar]
- Wang X., Yang C., 2016. Programmed cell death and clearance of cell corpses in Caenorhabditis elegans. Cell. Mol. Life Sci. 73: 2221–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang C., Chai J., Shi Y., Xue D., 2002. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science 298: 1587–1592. [DOI] [PubMed] [Google Scholar]
- Wang X., Wu Y. C., Fadok V. A., Lee M. C., Gengyo-Ando K., et al. , 2003. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 302: 1563–1566. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang J., Gengyo-Ando K., Gu L., Sun C. L., et al. , 2007. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat. Cell Biol. 9: 541–549. [DOI] [PubMed] [Google Scholar]
- Wang X., Li W., Zhao D., Liu B., Shi Y., et al. , 2010. Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat. Cell Biol. 12: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J., 1988 The anatomy, pp. 81–122 in The nematode Caenorhabditis elegans, Vol. 17, edited by W. B. Wood. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- White J. G., Southgate E., Thomson J. N., 1991. On the nature of undead cells in the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 331: 263–271. [Google Scholar]
- Winn J., Carter M., Avery L., Cameron S., 2011. Hox and a newly identified E2F co-repress cell death in Caenorhabditis elegans. Genetics 188: 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Wallen H. D., Nunez G., 1997. Interaction and regulation of subcellular localization of CED-4 by CED-9. Science 275: 1126–1129. [DOI] [PubMed] [Google Scholar]
- Wu W. S., Heinrichs S., Xu D., Garrison S. P., Zambetti G. P., et al. , 2005. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123: 641–653. [DOI] [PubMed] [Google Scholar]
- Wu Y. C., Horvitz H. R., 1998a The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93: 951–960. [DOI] [PubMed] [Google Scholar]
- Wu Y. C., Horvitz H. R., 1998b C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature 392: 501–504. [DOI] [PubMed] [Google Scholar]
- Wu Y. C., Stanfield G. M., Horvitz H. R., 2000. NUC-1, a caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 14: 536–548. [PMC free article] [PubMed] [Google Scholar]
- Wu Y. C., Tsai M. C., Cheng L. C., Chou C. J., Weng N. Y., 2001. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev. Cell 1: 491–502. [DOI] [PubMed] [Google Scholar]
- Wyllie A. H., 1980. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284: 555–556. [DOI] [PubMed] [Google Scholar]
- Xiao H., Chen D., Fang Z., Xu J., Sun X., et al. , 2009. Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol. Biol. Cell 20: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Liu Y., Zhao L., Gan Q., Wang X., et al. , 2014. The lysosomal cathepsin protease CPL-1 plays a leading role in phagosomal degradation of apoptotic cells in Caenorhabditis elegans. Mol. Biol. Cell 25: 2071–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D., Horvitz H. R., 1997. Caenorhabditis elegans CED-9 protein is a bifunctional cell-death inhibitor. Nature 390: 305–308. [DOI] [PubMed] [Google Scholar]
- Xue D., Shaham S., Horvitz H. R., 1996. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 10: 1073–1083. [DOI] [PubMed] [Google Scholar]
- Yan B., Memar N., Gallinger J., Conradt B., 2013. Coordination of cell proliferation and cell fate determination by CES-1 snail. PLoS Genet. 9: e1003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N., Gu L., Kokel D., Chai J., Li W., et al. , 2004. Structural, Biochemical, and Functional Analyses of CED-9 Recognition by the Proapoptotic Proteins EGL-1 and CED-4. Mol. Cell 15: 999–1006. [DOI] [PubMed] [Google Scholar]
- Yan N., Chai J., Lee E. S., Gu L., Liu Q., et al. , 2005. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature 437: 831–837. [DOI] [PubMed] [Google Scholar]
- Yang H., Chen Y. Z., Zhang Y., Wang X., Zhao X., et al. , 2015. A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat. Commun. 6: 5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P. T., Lorenowicz M. J., Silhankova M., Coudreuse D. Y., Betist M. C., et al. , 2008. Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev. Cell 14: 140–147. [DOI] [PubMed] [Google Scholar]
- Yang X., Chang H. Y., Baltimore D., 1998. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281: 1355–1357. [DOI] [PubMed] [Google Scholar]
- Yang Y., Lu J., Rovnak J., Quackenbush S. L., Lundquist E. A., 2006. SWAN-1, a Caenorhabditis elegans WD repeat protein of the AN11 family, is a negative regulator of Rac GTPase function. Genetics 174: 1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Lai H. J., Lin T. W., Chen C. S., Lo S. J., 2015. Loss of DNase II function in the gonad is associated with a higher expression of antimicrobial genes in Caenorhabditis elegans. Biochem. J. 470: 145–154. [DOI] [PubMed] [Google Scholar]
- Yu X., Lu N., Zhou Z., 2008. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 6: e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Odera S., Chuang C. H., Lu N., Zhou Z., 2006. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell 10: 743–757. [DOI] [PubMed] [Google Scholar]
- Yuan J., Horvitz H. R., 1992. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development 116: 309–320. [DOI] [PubMed] [Google Scholar]
- Yuan J., Shaham S., Ledoux S., Ellis H. M., Horvitz H. R., 1993. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75: 641–652. [DOI] [PubMed] [Google Scholar]
- Yuan J. Y., Horvitz H. R., 1990. The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev. Biol. 138: 33–41. [DOI] [PubMed] [Google Scholar]
- Zakharova L., Dadsetan S., Fomina A. F., 2009. Endogenous Jmjd6 gene product is expressed at the cell surface and regulates phagocytosis in immature monocyte-like activated THP-1 cells. J. Cell. Physiol. 221: 84–91. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Hodgkin J., 1992. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell 70: 237–249. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang H., Kage-Nakadai E., Mitani S., Wang X., 2012. C. elegans secreted lipid-binding protein NRF-5 mediates PS appearance on phagocytes for cell corpse engulfment. Curr. Biol. 22: 1276–1284. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Caron E., Hartwieg E., Hall A., Horvitz H. R., 2001a The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev. Cell 1: 477–489. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Hartwieg E., Horvitz H. R., 2001b CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104: 43–56. [DOI] [PubMed] [Google Scholar]
- Zou H., Henzel W. J., Liu X., Lutschg A., Wang X., 1997. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90: 405–413. [DOI] [PubMed] [Google Scholar]
- Zou H., Li Y., Liu X., Wang X., 1999. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274: 11549–11556. [DOI] [PubMed] [Google Scholar]
- Zou W., Lu Q., Zhao D., Li W., Mapes J., et al. , 2009. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 5: e1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullig S., Neukomm L. J., Jovanovic M., Charette S. J., Lyssenko N. N., et al. , 2007. Aminophospholipid translocase TAT-1 promotes phosphatidylserine exposure during C. elegans apoptosis. Curr. Biol. 17: 994–999. [DOI] [PubMed] [Google Scholar]