Abstract
Coilin is a marker protein for subnuclear organelles known as Cajal bodies, which are sites of various RNA metabolic processes including the biogenesis of spliceosomal small nuclear ribonucleoprotein particles. Through self-associations and interactions with other proteins and RNA, coilin provides a structural scaffold for Cajal body formation. However, despite a conspicuous presence in Cajal bodies, most coilin is dispersed in the nucleoplasm and expressed in cell types that lack these organelles. The molecular function of coilin, particularly of the substantial nucleoplasmic fraction, remains uncertain. We identified coilin loss-of-function mutations in a genetic screen for mutants showing either reduced or enhanced expression of an alternatively spliced GFP reporter gene in Arabidopsis thaliana. The coilin mutants feature enhanced GFP fluorescence and diminished Cajal bodies compared with wild-type plants. The amount of GFP protein is several-fold higher in the coilin mutants owing to elevated GFP transcript levels and more efficient splicing to produce a translatable GFP mRNA. Genome-wide RNA-sequencing data from two distinct coilin mutants revealed a small, shared subset of differentially expressed genes, many encoding stress-related proteins, and, unexpectedly, a trend toward increased splicing efficiency. These results suggest that coilin attenuates splicing and modulates transcription of a select group of genes. The transcriptional and splicing changes observed in coilin mutants are not accompanied by gross phenotypic abnormalities or dramatically altered stress responses, supporting a role for coilin in fine tuning gene expression. Our GFP reporter gene provides a sensitive monitor of coilin activity that will facilitate further investigations into the functions of this enigmatic protein.
Keywords: alternative splicing, Arabidopsis thaliana, Cajal body, coilin, stress
COILIN is an evolutionarily conserved protein with multiple proposed functions that are all connected to RNA-processing pathways (Bellini 2000; Hebert, 2010, 2013; Machyna et al., 2015). Coilin is best known as the signature protein of Cajal bodies (CBs), which are nonmembrane bound, nuclear suborganelles involved in various RNA metabolic processes, including biogenesis, maturation, and recycling of spliceosomal small nuclear ribonucleoprotein particles (snRNPs), histone mRNA processing, and telomere maintenance (Hebert, 2010, 2013; Machyna et al., 2015). CBs are dynamic entities that change in size and number depending on the cell type, cell cycle, developmental stage, and metabolic state (Boudonck et al., 1998, 1999). Spliceosomal snRNPs, which are composed of small nuclear RNAs (snRNAs) and snRNP-specific proteins, are indispensable for pre-mRNA splicing. Accordingly, CBs are prominent in cells with high transcriptional activity and pre-mRNA splicing requirements (Hebert 2010).
Twenty-five years after its discovery in an analysis of autoimmune patient sera (Andrade et al. 1991; Raska et al., 1991), coilin is still described as an “enigmatic” (Machyna et al. 2014) and a “tricky” (Machyna et al., 2015) protein. In all organisms studied so far, including Arabidopsis thaliana, coilin is critical for CB organization and integrity, presumably by serving as a scaffold for CB assembly (Collier et al., 2006; Hebert 2010; Machyna et al., 2015). Self-associations and interactions with RNA and other CB proteins are thought to underlie the ability of coilin to promote CB assembly; however, the mechanisms initiating CB formation are not yet fully understood (Hebert 2010; Rajendra et al., 2010; Makarov et al., 2013; Machyna et al., 2015; Novotný et al., 2015). Notably, despite its acknowledged role as a CB marker protein, most coilin in nuclei is not situated in CBs but is dispersed in the nucleoplasm (Lam et al., 2002). Moreover, coilin is constitutively expressed in most cell types, including those that do not contain visible CBs (Cioce and Lamond 2005; Hebert 2010). The molecular function of coilin, particularly of the sizeable nucleoplasmic portion, remains uncertain.
Even though coilin is not highly conserved at the amino acid sequence level, recognizable homologs have been identified in primitive and advanced metazoans and in plants, although not yet in Saccharomyces cerevisiae or Caenorhabditis elegans (Makarov et al., 2013; Machyna et al., 2015). A shared domain structure among animal and plant coilins has been proposed following an analysis of the secondary structure of Arabidopsis coilin (Makarov et al., 2013). The most conserved regions of coilin include a self-association domain at the N-terminus, which is required for CB formation, and an atypical Tudor-like domain at the C-terminal region (Shanbhag et al., 2010), which mediates interactions with several snRNP proteins (Hebert et al., 2001; Xu et al., 2005). The two conserved regions at the N- and C-termini flank a central highly disordered region (Makarov et al., 2013). Although coilin lacks conventional RNA binding motifs, several degenerate RNA recognition motifs were identified in the N-terminal region and central disordered region (Makarov et al., 2013; Machyna et al., 2015). In animal cells, coilin interacts with many nuclear small noncoding RNA species, including small Cajal body-specific RNAs (scaRNAs), which guide modification of the snRNA component of snRNPs (Enwerem et al., 2014), as well as snRNAs, snoRNAs, and telomerase RNAs (Machyna et al., 2014). Coilin has been reported to bind to double-stranded DNA (Broome and Hebert 2012) and to associate with genes encoding snRNAs, snoRNAs, and histones (Hebert 2010; Machyna et al., 2015), suggesting that it may influence transcription, maturation of transcripts, or higher-order chromatin structure (Machyna et al., 2015).
Findings regarding the necessity of coilin for development vary depending on the organism under investigation. Coilin-deficient mutants in Arabidopsis (Collier et al., 2006) and Drosophila (Liu et al., 2009) show no striking developmental phenotypes, whereas coilin depletion in zebrafish (Strzelecka et al., 2010) and mice (Walker et al., 2009) results in embryonic lethality and semilethality, respectively. These discrepant requirements for coilin may reflect distinct developmental mechanisms or variations in pre-mRNA splicing demands among different organisms (Machyna et al., 2015). The viability of coilin mutants of Arabidopsis and Drosophila demonstrates that CBs are not absolutely required for snRNP assembly and activity but are likely to promote the efficiency of these processes (Rajendra et al., 2010).
As a prominent component of CBs and a direct interactor with several snRNP proteins and snRNAs (Hebert et al., 2001; Xu et al., 2005; Enwerem et al., 2014; Machyna et al., 2014), coilin has been implicated in pre-mRNA splicing. Findings available so far indicate that coilin deficiencies have a negative impact on splicing. Coilin knockdown by RNAi in HeLa cells reduced splicing efficiency of an artificial reporter gene (Whittom et al., 2008). Zebrafish depleted of coilin using a morpholino approach exhibited splicing defects that could be alleviated by injection of fully assembled snRNPs, supporting the idea that coilin helps to concentrate snRNP components in CBs and facilitate snRNP assembly (Strzelecka et al., 2010). A full understanding of the roles of coilin in snRNP metabolism and pre-mRNA splicing awaits further investigation. To our knowledge, genome-wide studies of pre-mRNA splicing efficiency in coilin loss-of-function mutants, which would reveal the global consequences of a complete coilin deficit on splicing, have not yet been reported for any organism.
We are using genetic approaches to identify new factors that influence pre-mRNA splicing efficiency and alternative splicing in plants (Reddy et al., 2013; Meyer et al., 2015). For this, we exploit a transgenic Arabidopsis line containing an alternatively spliced GFP reporter gene under the control of viral transcriptional regulatory elements (Sasaki et al., 2015). Of three possible splice variants, only a “short” transcript arising from the splicing of a U2-type intron with noncanonical AT-AC splice sites (Sharp and Burge 1997) gives rise to a translatable GFP mRNA (Sasaki et al., 2015). In a previous genetic suppressor screen based on the GFP reporter gene, two mutants showing reduced GFP expression were found to be defective, respectively, in the core spliceosomal protein PRP8 (pre-mRNA processing 8) and in a novel, conserved protein termed RTF2 (replication termination factor 2), which may be involved in ubiquitin-based regulation of the spliceosome (Sasaki et al., 2015). In both the prp8 and rtf2 mutants, splicing of the AT-AC intron was impaired, resulting in decreased levels of translatable GFP mRNA and enhanced accumulation of an unspliced, untranslatable GFP transcript (Sasaki et al., 2015). An analysis of genome-wide RNA-sequencing (RNA-seq) data indicated that ∼16% of introns are less efficiently spliced in the two mutants (Sasaki et al., 2015).
The identification of the prp8 and rtf2 mutants in our suppressor screen validated the use of the alternatively spliced GFP reporter gene system for identifying both core spliceosomal proteins and novel putative regulators of splicing. Here we describe the identification in a modified screen of a new complementation group displaying enhanced GFP expression, which was found to be attributable to loss-of-function mutations in the gene encoding coilin. Our analysis of genome-wide RNA-seq data from the mutants indicates that coilin modulates transcript levels from a small subset of genes, many encoding stress-related factors, and can attenuate splicing efficiency, a result that contrasts with previous studies. We suggest that coilin acts at multiple levels to fine tune the expression of a select group of genes that may contribute to environmental adaptation.
Materials and Methods
Plant materials and forward genetic screen
This study used a transgenic Arabidopsis line (ecotype Columbia, Col) that is homozygous for a target (T) locus containing an alternatively-spliced GFP reporter gene, which is expressed in meristem regions at the shoot and root apices and in the hypocotyl (stem) of young seedlings (Kanno et al., 2008; Sasaki et al., 2015). In the absence of a “silencer” locus (Kanno et al., 2008; Sasaki et al., 2015), the GFP reporter gene has been continuously expressed in the T line for ∼10 years, and thus provides a suitably stable system for use in forward genetic screens. Owing to negligible amounts of GFP small RNAs (Supplemental Material, Figure S1) and little or no DNA methylation in the upstream promoter–enhancer region (Sasaki et al., 2014), the intermediate GFP expression level in the T line is unlikely to result from either canonical posttranscriptional gene silencing or DNA methylation-mediated transcriptional gene silencing. This supports the hypothesis that a balanced ratio of alternatively spliced transcripts maintains moderate levels of GFP translatable mRNA (Figure 1). For simplicity, we refer to the nonmutagenized T line as “wild-type” in this paper.
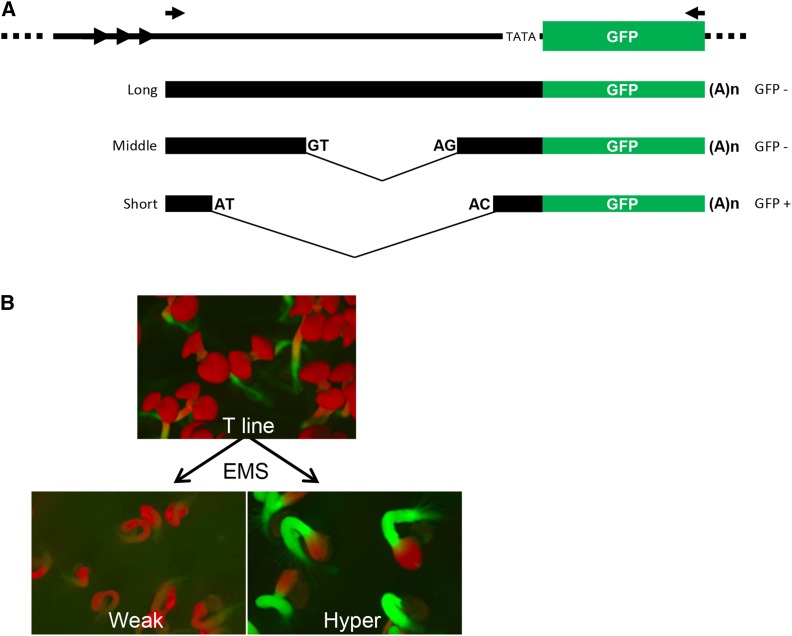
Figure 1.
GFP reporter gene system and GFP phenotypes in mutants. (A) The GFP reporter gene in the T line and alternative splicing of GFP pre-mRNA. The GFP coding region (green bar) is under the control of virus-derived transcriptional regulatory elements: a truncated 35S promoter (TATA) and the endogenous pararetrovirus (EPRV) enhancer (∼1.2 kb, black bar), which contains a tandem repeat (three copies of a 41–42-bp monomer, arrowheads) in the 5′ distal portion. Transcription of GPF pre-mRNA begins around the tandem repeat region. Two opposing arrows above the diagram indicate the positions of primers used for RT-PCR to detect three major GFP transcripts: the untranslatable “long” and “middle” transcripts, and a “short” translatable mRNA. The middle and short transcripts result from alternative splicing of U2-type introns containing canonical (GT-AG) and noncanonical (AT-AC) splice sites, respectively (Sasaki et al., 2015). (B) GFP phenotypes in seedlings. The wild-type T line shows an intermediate level of GFP fluorescence visible mainly in the stem (hypocotyl) and shoot and root apices of young seedlings. Mutants obtained following EMS mutagenesis of the T line could display either reduced (“Weak”) or stronger (“Hyper”) GFP fluorescence. Cotyledons (the first set of leaves sprouting from the seed) appear red owing to auto-fluorescence of chlorophyll at the excitation wavelength for GFP.
To carry out a forward genetic screen to identify splicing factors, ∼40,000 seeds (M1 generation) of the wild-type T line were treated with the chemical mutagen ethyl methanesulfonate (EMS) according to a standard protocol (Kim et al., 2006). The M1 seeds were sown on soil and allowed to flower and self-fertilize to produce M2 seeds, which represent the first generation when recessive mutations can be homozygous and exhibit a phenotype. Approximately 280,000 1–2-week-old M2 seedlings (about seven M2 progeny from each M1 plant) (Haughn and Somerville 1990) cultivated under sterile conditions on solid Murashige and Skoog (MS) medium in square Petri dishes were screened for GFP fluorescence under a fluorescence stereo microscope. Seedlings displaying a GFP-weak or Hyper-GFP phenotype were among those selected for additional analysis, which included sequencing the GFP reporter gene to confirm a wild-type sequence for the GFP coding and upstream regions. The focus in this paper is on the hyper-gfp1 (hgf1) complementation group.
Next-generation mapping (NGM)
NGM was used to determine the causal mutation in two members of the hgf1 complementation group. For NGM, an hgf1 mutant was crossed with Arabidopsis ecotype Landsberg erecta (Ler) to produce F1 plants, which were allowed to self-fertilize to produce F2 seeds. The F2 seeds were sterilized and sown on solid MS medium. F2 seedlings showing a Hyper-GFP phenotype were selected for DNA isolation. Pooled DNA from at least 50 Hyper-GFP F2 seedlings was used for sequencing on an Illumina platform and analyzed for the position of the causal mutation according to a published NGM protocol (Austin et al., 2011, 2014).
DNA sequence analysis of GFP and coilin genes
Primers used for sequencing the GFP reporter gene, the EPRV upstream sequence, and coilin (At1g13030) are shown in Table S1.
Complementation test
A construct encoding a coilin-DsRed-Monomer fusion protein under the control of the endogenous coilin promoter (367 bp upstream of the ATG start codon) and rbcS3C transcription terminator (Benfey et al., 1989) was inserted into the binary vector pPZP221, which encodes resistance to gentamicin (Hajdukiewicz et al., 1994). The modified binary vector was transferred into Agrobacterium tumefaciens strain ASE, which was used to transform the coilin mutant hgf1-8 (P439L) using the floral dip procedure (Clough and Bent 1998). T1 seedlings were selected on solid MS medium containing gentamicin and later transferred to soil. T2 seeds resulting from self-fertilization of T1 plants were sown on gentamicin-containing MS medium and scored for segregation of gentamicin resistance and GFP fluorescence. Complementation of the hgf1-8 mutation was considered successful if an intermediate level of GFP fluorescence similar to that observed in the wild-type T line was restored in gentamicin-resistant seedlings.
Effect of coilin mutation on CB integrity
A coilin mutant homozygous for the hgf1-7 mutation (W437*), which can be genotyped using a cleaved amplified polymorphic sequences (CAPS) marker, was crossed to a transgenic line expressing U2B″:GFP, which is a CB-specific fluorescent marker (Collier et al., 2006). The resulting F1 plants were allowed to self-fertilize to produce F2 seeds, which were germinated on solid MS medium containing phosphinothricin (PPT). F2 seedlings were selected for PPT resistance (contributed by the U2B″:GFP line) and a GFP-negative phenotype in the hypocotyl and meristem regions, indicating absence of the T construct. Seedlings selected in this way were transferred to soil and later genotyped for the homozygous hgf1-7 mutation using the primers shown in Table S1. The presence of CBs in leaf nuclei of wild-type and homozygous hgf1-7 plants containing the U2B″:GFP gene was assessed using a TCS LSI-III Confocal Microscope System. At least 20 leaf nuclei were examined for each genotype.
Western blots
Western blotting to detect GFP protein was carried out using protein extracts isolated from 2-week-old mutant and wild-type seedlings, as described previously (Fu et al., 2015).
RT-PCR of GFP RNAs
Semiquantitative RT-PCR to detect GFP RNAs was performed using total RNA isolated from 2-week-old seedlings according to a previously published protocol (Sasaki et al., 2015). GFP and actin primers are shown in Table S1.
RNA-seq
Total RNA was isolated from 2-week-old seedlings of the wild-type T line and from two coilin mutants: hgf1-1 (R40*) and hgf1-8 (P439L). Because the two mutant lines contain loss-of-function mutations in the same gene (coilin) and were derived from the same T line, they were regarded as “mutant” replicates for the purposes of this study. Library preparation and RNA-seq were carried out as described previously (Sasaki et al., 2015). Two sequencing experiments were performed: in one, a total of 305 million reads were sequenced for technical triplicates (same library sequenced three times) of the wild-type T line and hgf1-8 and, in the second, a total of 173 million reads were sequenced for technical duplicates (two different libraries sequenced) of the T line and hgf1-1.
RNA-seq reads were mapped in two stages. In the first stage, the reads were mapped to the TAIR10 transcriptome using Bowtie 2 (Langmead and Salzberg 2012). Only read pairs that had been both mapped to the same transcript(s) were kept, and their alignments were translated to the TAIR10 genome. In the second stage, rest reads were mapped to the TAIR10 genome using BLAT (Kent 2002) with the default setting. Only best alignments with an identity of no less than 90% were accepted for computation and >95% of reads were accepted for every replicate (see Table S2 for mapping statistics). RackJ (http://rackj.sourceforge.net) was then used to compute the read counts of all genes and average the depths of all exons and all introns.
The read counts of all samples were normalized using the trimmed mean of M values method (Robinson and Oshlack 2010) and transformed into logarithmic counts per million (logCPM) using the voom method (Law et al., 2014) with parameter normalize = “none”. Adjusted reads per kilobase model (RPKM) values were computed based on logCPMs and used for Z-tests as described in (Lan et al., 2013). In this study, we defined a gene as differentially expressed if its P-value by the Z-test was ≤0.01.
The preference of intron retention events was measured using a χ2 test for goodness-of-fit as described by Sasaki et al. (2015), where read depths of an intron in two samples were compared with the background of read depths of neighboring exons. In so doing, the underlying null hypothesis assumes that the chances for an intron to be retained are the same in the two samples, and a significant P-value indicates that the chance of intron retention was altered in one of the two samples. Given an intron with a P-value of ≤0.01, we defined it as more efficiently spliced if the ratio intron_depth/exon_depth in the mutant was smaller than that in the control; otherwise, we defined it as an intron of increased retention.
Abiotic stress tests: salt and chilling
We tested four coilin mutants for sensitivity to salt stress by germinating and growing seedlings on sterile MS medium containing either no additional NaCl, 100 mM NaCl, or 150 mM NaCl (Verslues et al., 2006; Ito et al., 2016). The seedlings were incubated at 23° under a 16-hr light:8-hr dark cycle, and observed over a 6-week period for growth defects of the root and shoot portions of the seedling. Chilling sensitivity was monitored by germinating seedlings on duplicate plates containing solid MS medium and incubating at 23° under a 16-hr light:8-hr dark cycle for 2 weeks. Half of the plates were then placed at 5° under the same light:dark regime for 4 weeks, after which they were returned to 23°. The growth and appearance of chilled and control seedlings were then compared at daily intervals for 4 weeks.
Virus infection
Severe (TuGR) and mild (TuGK) strains of Turnip mosaic virus (TuMV) (Kung et al., 2014), and Cucumber mosaic virus Fny strain (CMV) were maintained on Nicotiana benthamiana plants. For challenge inoculation, 2.5-week-old Arabidopsis seedlings (four coilin mutants and wild-type T line) were mechanically inoculated with TuGR, TuGK, or CMV, which were prepared from 0.5 g of infected N. benthamiana leaves using 2 ml of 0.01 M sodium phosphate buffer (pH 7.2). At 7 days after the virus inoculation, the virus accumulation was evaluated by examining the development of symptoms in the plants. Presented viruses were monitored by indirect ELISA as follows: 0.5 g of infected tissues were extracted with 5 ml of coating buffer (15 mM Na2CO3, 34.9 mM NaHCO3, 3 mM NaN3, pH 9.6), and then the extract was diluted 40× for indirect ELISA assay with anti-TuMV CP antiserum for TuGR or TuGK detection, or anti-CMV CP antiserum for CMV detestation (Niu et al., 2006). The results were recoded as absorbance at 405 nm using a VersaMax Tunable Microplate Reader (Molecular Devices, Sunnyvale, CA).
Data availability
Seeds of the wild-type T line and the coilin mutants will be deposited at the Arabidopsis Biological Resource Center, and are currently available on request from Matzke’s laboratory. RNA-seq data are available from NCBI SRA under accession number SRP071829.
Results
Forward screen and identification of hgf1 (coilin) mutants
A schematic drawing of the alternatively spliced GFP reporter gene in the T line is shown in Figure 1A. Three major transcripts issue from the GFP gene, but only the short transcript corresponds to a translatable GFP mRNA (Sasaki et al., 2015). For the genetic screen to identify splicing factors, seeds of the homozygous T line were treated with EMS and sown on soil to produce the M1 generation. M2 seeds obtained from the self-fertilization of the M1 plants (M2 is the first generation when a recessive mutation can be homozygous and show a phenotype) were harvested, and a sampling of M2 seeds from each M1 parent was sown on solid MS medium. After germination, the M2 seedlings were scored for GFP fluorescence by visualization under a stereo fluorescence microscope.
The M2 seedling population contained an assortment of putative mutants displaying different GFP phenotypes. GFP-negative mutants were found to harbor loss-of-function mutations in the GFP coding sequence (Fu et al., 2015), and are not considered further here. We also observed mutants in which expression of the GFP reporter gene was either weaker (“GFP-weak”) or stronger (“Hyper-GFP”) than in wild-type plants (Figure 1B). Although the GFP-weak phenotype was expected from previous studies (Sasaki et al., 2015), the Hyper-GFP phenotype was new and unanticipated. The bidirectional expression changes observed in the various mutants indicated that GFP is normally expressed at an intermediate level in wild-type plants, presumably owing to a stable balance of the three alternatively spliced transcripts. Based on our earlier findings (Sasaki et al., 2015), we hypothesize that the GFP-weak and Hyper-GFP mutant phenotypes reflect mutations that alter the ratios of the three alternatively spliced GFP transcripts, resulting in either reduced or increased amounts of the short translatable GFP mRNA, respectively.
Approximately 50 Hyper-GFP mutants were identified in the M2 seedling population. Progeny from intercrosses among these mutants revealed at least five hyper-gfp (hgf) complementation groups, the largest of which, hgf1, contained 14 members. We confirmed by sequence analysis that the GFP coding sequence and upstream promoter–enhancer region were not mutated in the hgf1 mutants, indicating that the Hyper-GFP phenotype was likely due to mutations in a gene required for maintaining intermediate GFP gene expression. NGM (Austin et al., 2011, 2014) of two hgf1 mutants revealed independent point mutations in the gene encoding coilin (At1g13030), which is present in a single copy in Arabidopsis. Sequencing of the coilin gene in the remaining 12 hgf1 mutants identified further independent point mutations, adding up to a total of eight different hgf1 alleles: hgf1-1 to hgf1-8 (Figure 2 and Figure S2). The eight hgf1 alleles are distinct from three other coilin alleles, ncb-1, ncb-2, and ncb-3 (no cajal bodies) (Figure S3), which were identified in a prior screen for Arabidopsis mutants with altered CBs (Collier et al., 2006). Four hgf1 mutations (hfg1-1, hgf1-5, hgf1-6, and hgf1-7) create premature termination codons, and two (hfg1-2 and hgf1-4) destroy splice sites. Two mutations result in amino acid substitutions: E76K (hgf1-3), which is in the self-association domain at the N-terminus, and P439L (hgf1-8), which is in the Tudor-like domain in the C-terminal half of the protein (Figure 2 and Figure S3). E76 is conserved in several plant species (Figure S3) while P439 is conserved in plant and animal coilins (Figure S3 and Figure S4). Coilin is ubiquitously expressed in Arabidopsis, and is particularly highly expressed in the shoot apex and flower buds (Arabidopsis eFP Browser; Winter et al., 2007).
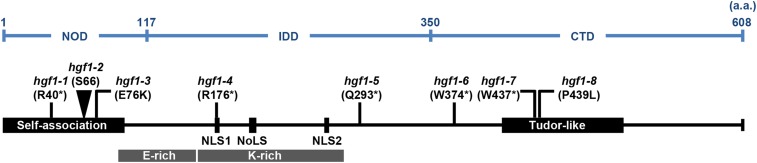
Figure 2.
Domain organization of Arabidopsis coilin and the positions of hgf loss-of-function mutations. Arabidopsis coilin consists of 608 amino acids. The N-terminal self-association domain and C-terminal Tudor-like domain are the most highly conserved regions of coilin proteins. Analysis of the secondary structure of Arabidopsis coilin predicted three domains: the N-terminal globular domain (NOD), the internal disordered domain (IDD), and the C-terminal domain (CTD) (Makarov et al., 2013). Also shown are two nuclear localization (NLS) signals and one cryptic nucleolar localization signal (NoLS) as well as E-rich and K-rich domains (Makarov et al., 2013). The eight hgf mutations retrieved in our forward genetic screen are indicated. Some alleles (hfg1-1, hgf1-3, hgf1-6, and hgf1-8) were obtained more than once. The corresponding nucleotide changes are shown in Figure S2.
The eight hgf1 alleles are all recessive, as indicated by the restoration of an intermediate GFP phenotype observed in the wild-type T line in F1 progeny generated by crossing the coilin mutants with the parental T line (data not shown). The intermediate GFP phenotype of the wild-type T line was also restored after introducing a wild-type coilin coding sequence under the control of the endogenous coilin promoter into the hgf1-8 mutant (Figure S5). Together with the finding of multiple coilin alleles, the complementation test confirmed that the hgf1 mutations in the coilin gene were responsible for the Hyper-GFP phenotype. Apart from a short delay in flowering, the coilin mutants did not show any consistent growth, developmental, or reproductive phenotypes, a result that is in accord with previous findings (Collier et al., 2006). As expected, CBs dissipated in plants containing a coilin mutation (Figure 3).
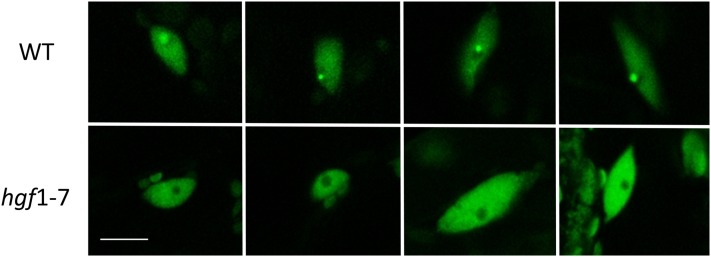
Figure 3.
Dispersion of CBs in a coilin mutant. Owing to expression of the CB marker U2B″:GFP, CBs are visible as single, highly fluorescent spots, often close to the nucleolus (dark spherical area), in leaf nuclei of wild-type plants (top). By contrast, in the hgf1-7 coilin mutant, CBs are either uniformly absent or much smaller and less intensely fluorescent in all cells examined. The white bar indicates 10 µm.
Effects of coilin mutations on GFP protein and GFP pre-mRNA splicing
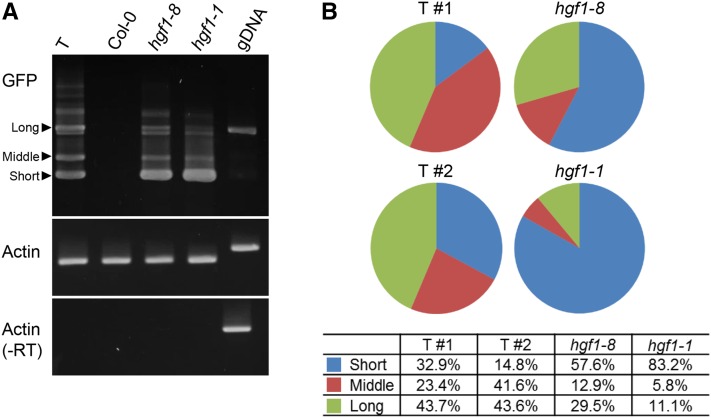
Relative to the wild-type T line, the eight hgf1 alleles condition a clear Hyper-GFP phenotype that can be visualized in seedlings (Figure 4A). To estimate the levels of GFP protein in the coilin mutants, we performed Western blots using a GFP antibody. The amount of GFP protein detected using this technique was several-fold higher in the coilin mutants compared with the wild-type T line (Figure 4B), confirming that the enhanced GFP fluorescence was due to increased levels of GFP protein. Two coilin alleles, hgf1-1 (R40*) and hgf1-8 (P439L), were selected for RNA analysis. Semiquantitative RT-PCR was used to detect the three alternatively spliced GFP transcripts in the mutants and wild-type plants. Compared with the wild-type T line, the coilin mutants contained increased levels of the short translatable GFP transcript and decreased levels of the untranslatable middle and long transcripts (Figure 5A). These results suggested more-efficient splicing of the U2-type intron (with noncanonical AT-AC splice sites) in the coilin mutants.
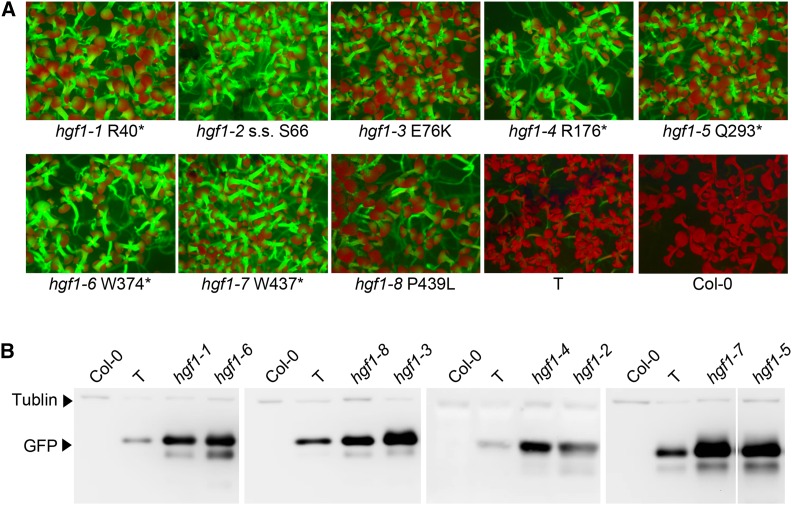
Figure 4.
Levels of GFP protein in coilin mutants. (A) Hyper-GFP fluorescence in coilin mutant seedlings. Appearance of ∼1–2-week-old seedlings of coilin mutants hgf1-1 through hgf1-8 as well as the wild-type T line and untransformed Col-0 growing on solid MS medium as visualized under a fluorescence stereo microscope. GFP fluorescence is high in the hypocotyls. Cotyledons appear red owing to auto-fluorescence of chlorophyll at the excitation wavelength of GFP. (B) Western blot analysis of GFP protein in coilin mutants. Separate lanes for the wild-type T line and nontransgenic Col-0 lanes are shown for samples that were run on separate gels. A tubulin loading control is visible at the top of each lane. The second smaller band migrating slightly below the GFP protein is likely a degradation product that is particularly noticeable when the GFP protein levels are high.
Figure 5.
Abundance of GFP RNA isoforms in coilin mutants. (A) RT-PCR of GFP RNAs in coilin mutants. Semiquantitative RT-PCR was used to assess the accumulation of long, middle, and short GFP transcripts in two coilin mutants (hgf1-1 and hgf1-8), the wild-type T line, and nontransgenic Col-0. Actin is shown as a constitutively expressed control. –RT, no reverse transcriptase; gDNA, genomic DNA. (B) Percentages of the three major splice variants of GFP RNA were predicted based on RNA-seq data (Table S5, Table S8, and Table S9).
RNA-seq analysis of transcription and splicing efficiency
To examine transcription and pre-mRNA splicing in more detail, we carried out RNA-seq on RNA isolated from hgf1-1, hgf1-8, and the wild-type T line. The RNA-seq data were analyzed for differentially expressed genes (DEGs) and splicing efficiency genome-wide as well as for differential transcription and splicing of the GFP reporter gene. To make a valid assessment of the consequences arising specifically from a coilin deficiency, we focused on findings that were shared between the two coilin mutants. Although the lack of a strict biological replication complicates the ability to perform quantitative analyses, the focus on results found in both mutant lines helps to provide robust results.
Using a cut-off of P < 0.01, steady state levels of GFP transcript increased to some extent in the two coilin mutants [∼1.6-fold in hgf1-8 (Table S3) and ∼1.1-fold in hgf1-1 (Table S4)]. Consistent with the semiquantitative RT-PCR experiment (Figure 5A), the RNA-seq data also revealed that alternative splicing of GFP pre-mRNA was altered in the coilin mutants (Table S5), with the percentage of short translatable transcript increasing from an average of ∼24% of the total GFP transcripts in the wild-type T line to 57.6% and 83.2%, respectively, in the hgf1-8 and hgf1-1 mutants (Figure 5B). These increases, which were accompanied by corresponding decreases in the percentages of untranslatable middle and long RNAs, represent fold changes of ∼1.9 (hgf1-8) and ∼3.2 (hgf1-1) in the level of the short translatable GFP transcript. The several-fold increases in GFP protein in the coilin mutants (Figure 4B) can thus be accounted for by both increases in steady state levels of the GFP transcript (∼1.1–1.6-fold) and more efficient splicing to produce enhanced (∼1.9–3.2-fold) quantities of the short translatable GFP mRNA.
Genome-wide transcription and splicing in coilin mutants
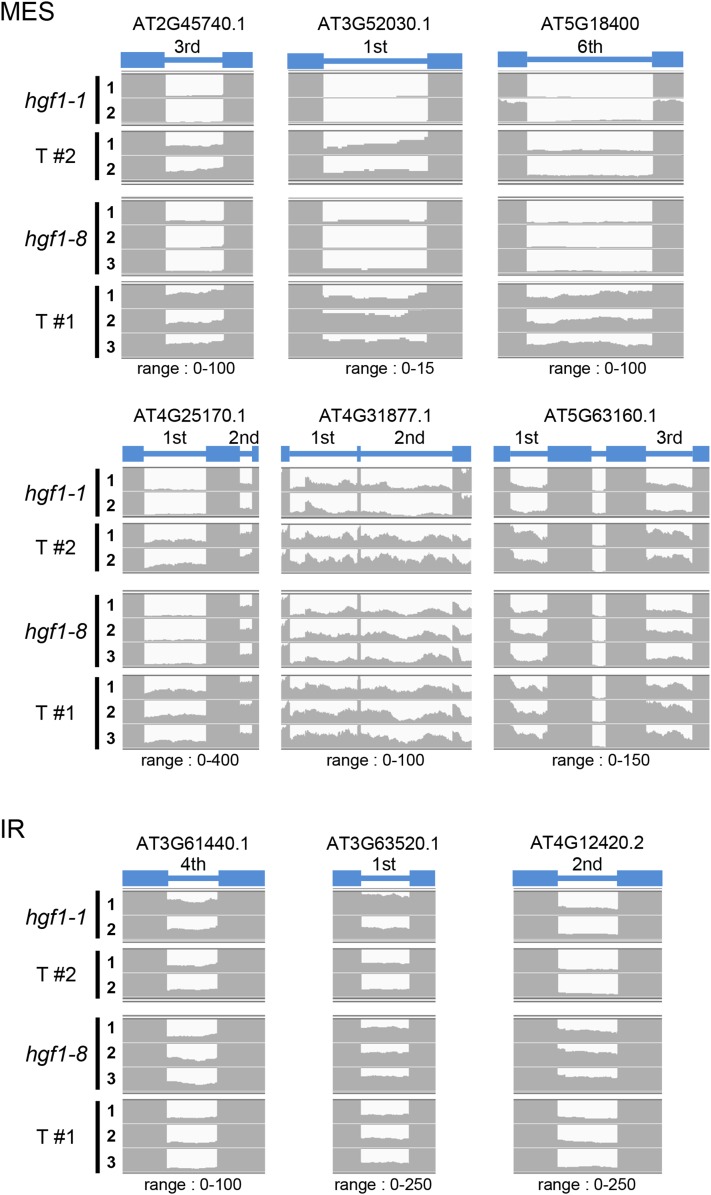
Analysis of the RNA-seq data revealed minor changes in transcription genome-wide in the coilin mutants. Out of 33,602 annotated Arabidopsis genes, only 102 DEGs (0.3% of the total genes) were shared between hgf1-1 and hgf1-8 (51 “UP”, 51 “DOWN”) (Table 1 and Table S6; the complete set of data on the DEGs for each mutant is given in Table S3 and Table S4, respectively). Given the proposed role of coilin in spliceosomal snRNP biogenesis, the consequences of the coilin mutations on splicing genome-wide were surprisingly limited, and tended to favor more-efficient splicing. From the total number of introns in Arabidopsis, only 381 (0.3% of the total introns) were affected in both coilin mutants. The 381 shared splicing changes comprised 356 more efficiently spliced (MES) introns and only 25 instances of increased intron retention (IR) (Table 1 and Table S7; the complete set of data on the MES and IR for each mutants is given in Table S8 and Table S9, respectively). There was no correlation between MES introns and the occurrence of alternative splicing or intron number, and nearly all cases concerned U2-type introns with canonical GT-AG splice sites (Table S7). The 356 MES events included multiple introns in 51 genes, indicating relatively consistent enhancement of splicing along a given pre-mRNA [Table S7 (Share_MES)]. By contrast, of the 25 IR events, multiple introns were only observed for one gene, suggesting more sporadic inefficiency of splicing [Table S7 (Share_IR)]. The results for representative genes showing either decreased or increased numbers of reads in introns (corresponding to MES and IR, respectively) in the coilin mutants compared with the wild-type T line are displayed schematically in Figure 6.
Table 1. Numbers of DEGs and differentially spliced introns in two coilin mutants.
| Number of DEGs | Number of introns affected | |||||
|---|---|---|---|---|---|---|
| hgf1-1 | hgf1-8 | hgf1-1 | hgf1-8 | Shared | ||
| Total number of genes | ||||||
| 33,602 | Up | 333 (0.99%) | 216 (0.64%) | 51 (0.15%) | ||
| Down | 255 (0.76%) | 348 (1.04%) | 51 (0.15%) | |||
| Total number of introns | ||||||
| 120,998 | IR | 1541 (1.27%) | 281 (0.23%) | 25 (0.02%) | ||
| MES | 1046 (0.86%) | 2163 (1.79%) | 356 (0.29%) | |||
The total gene number (33,602) includes nuclear genes, transposable element genes and pseudogenes counted directly from the annotation file of TAIR10 and excludes chloroplast and mitochondrial genes. The total intron number (120,998) was counted from merged gene models of these 33,602 genes. The numbers of up- and down-regulated DEGs and the numbers of IR or MES events in the single coilin mutants are shown as well as those that are shared between the two mutants (percentages of the respective total number in parentheses). The overlap between DEGs and MES/IR events is minor, with <10 genes in each category [DEGs UP/MES (three genes), DEGs DOWN/MES (five genes); DEGs UP/IR (six genes), DEGs DOWN/IR (no genes)] [Table S7 (+DEG)]. The “shared” category is presumed to reflect changes due specifically to a coilin deficiency. Full data sets are given in Table S3, Table S4, Table S6, Table S7, Table S8, and Table S9.
Figure 6.
Examples of introns affected in splicing efficiency in coilin mutants. The number of reads for several introns that show either more-efficient splicing (MES; upper part) or increased intron retention (IR; lower part) in coilin mutants containing either the hgf1-8 or hgf1-1 allele compared with the wild-type T line are visualized by the Integrative Genomic Viewer. The target intron and the exons before and after the intron are shown by the blue bars and blue boxes, respectively. The Arabidopsis Genome Initiative (AGI) number for the target intron-containing gene and the range for counting the reads are shown at the top and bottom of each figure. The numbers on the left side of each figure indicate the number of technical replicates for each indicated plant line. The genes shown here are not among those that are differentially expressed genes in the T and coilin mutant lines.
Stress-related genes represented in shared DEGs and shared MES events
Among the shared DEGs, a number of genes related to stress responses and senescence were identified. Of the 51 down-regulated genes, 13 have functions in photosynthesis, including multiple genes encoding chlorophyll A/B binding proteins, light-harvesting complexes, and photosystem I subunits [Table S6 (Share DOWN)]. Down-regulation of photosynthetic genes is a hallmark of senescing leaves (Breeze et al., 2011). Additional stress or defense-related genes were also down-regulated, including two defense-related WRKY transcription factors (WRKY33 and WRKY40) as well as salt and wound-inducible genes [Table S6 (Share DOWN)]. The up-regulated category also contained many stress- and senescence-related genes, including several metallothioneins and other metal-binding proteins, which are up-regulated during senescence (Breeze et al., 2011), and multiple genes encoding stress-responsive amino acid permeases and cold-acclimation factors [Table S6 (Share UP)]. Strikingly, ACC oxidase 2 (ACO2) and the transcription factor NAC83 were also among the up-regulated DEGs, and both were previously identified as senescence-enhanced genes [Table S6 (Share UP)] (Breeze et al., 2011). Overall, these results suggest a partial triggering of a premature senescence program, which provides a protective function to stressed plants (Breeze et al., 2011), in the coilin mutants.
Similarly to the shared DEG list, the shared MES category contains a number of genes generally involved in abiotic stress tolerance, disease resistance, and hormone responses [Table S7 (Share MES)]. However, the overlap between DEGs and MES/IR events was small, with <10 genes in each category [DEGs UP/MES (three genes), DEGs DOWN/MES (five genes); DEGs UP/IR (six genes), DEGs DOWN/IR (no genes)] [Table S7 (+DEG)].
Stress tests
The identification of stress- and senescence-related genes among the DEGs and MES introns prompted us to evaluate the response of coilin mutants to various stress treatments. We tested four different coilin mutants for sensitivity to salt and chilling stress (Verslues et al., 2006) and to RNA virus infection (Shaw et al., 2014). The four alleles tested included two amino acid substitutions, hgf1-3 (E76K) and hgf1-8 (P439L); a splice site mutation, hgf1-2 (S66); and a premature termination codon, hgf1-7 (W437*). We did not observe any consistent changes in the responses of the four coilin mutants to either salt or chilling treatment, which would be expected if any positive or negative effects were due specifically to the coilin deficiencies (data not shown) or to virus infection (Figure S6).
Discussion
We identified eight independent loss-of-function mutations in the Arabidopsis gene encoding coilin in a forward genetic screen based on an alternatively spliced GFP reporter gene under the control of virus-derived promoter–enhancer elements. The coilin mutations, which are all recessive, condition a Hyper-GFP phenotype relative to the wild-type T line used for mutagenesis. The Hyper-GFP phenotype reflects a several-fold increase in GFP protein levels that results from moderately elevated levels of GFP steady state transcripts as well as enhanced splicing to produce a translatable GFP mRNA. Whether the additive effect of increased transcript levels and splicing efficiency can account fully for the Hyper-GFP phenotype in coilin mutants is not yet known, and alterations in other processes, including post-transcriptional processing steps, nuclear envelope transit, or translation of GFP mRNA into protein, cannot presently be ruled out.
Based on current knowledge of coilin function, it is not clear why we retrieved coilin mutations in our screen. The proposed role of coilin in spliceosomal snRNP biogenesis would predict that coilin deficiencies should impair pre-mRNA splicing. Indeed, coilin depletion in animal systems has previously been associated with splicing defects (Whittom et al., 2008; Strzelecka et al., 2010). Yet, we observed enhanced splicing of a U2-type intron (with cryptic AT-AC splice sites) in GFP pre-mRNA. Similarly, even though the effects of coilin mutations on splicing genome-wide were very minor (only 0.3% of introns were affected in both coilin mutants), the trend was clearly toward more efficient splicing and not increased intron retention. This was particularly convincing for the 51 genes in which multiple introns were more efficiently spliced, consistent with a persistent enhancement of splicing along the pre-mRNA. Although GFP transcript levels were elevated in the coilin mutants, the effect of coilin deficiency on transcription genome-wide was very modest (affecting only 0.3% of the total genes) and included approximately equal numbers of up- and down-regulated genes. In HeLa cells, coilin knockdown reduced transcription rates and cell doubling times, indicating a negative effect of coilin depletion on transcription in addition to the above-mentioned reduction of splicing efficiency (Whittom et al., 2008). In principle, coilin could help to coordinate the regulation of transcription and splicing, which is considered a largely cotranscriptional process (Herzel and Neugebauer 2015). In our coilin mutants, however, there was only a slight overlap between differentially expressed genes and genes undergoing differential splicing events. These results suggest that coilin can independently influence either transcription or splicing of different target genes by mechanisms that remain to be determined. Understanding the ways in which coilin might in some cases coordinately modulate both transcription and splicing of the same gene also awaits further investigation.
To our knowledge, the only other forward genetic screen that identified mutations in coilin was carried out in Arabidopsis and involved screening for alterations in CB size and number (Collier et al., 2006). Thus, some special feature or combination of features of our GFP reporter gene system makes it particularly responsive to mutations in the coilin gene. Perhaps the extensive viral nature of the transcriptional regulatory elements upstream of the GFP reporter gene provides in some way a target for coilin. Connections between coilin function and virus infection have been made previously in plants (Shaw et al., 2014) and animals (James et al., 2010). The ∼1.2-kb enhancer region upstream of the GFP reporter gene is derived from a tobacco EPRV; Gregor et al., 2004) and the 54-bp minimal promoter is a truncated form of the 35S promoter of the cauliflower mosaic (pararetro)virus (CaMV) (Benfey et al., 1989). Initiation of GFP pre-mRNA transcription is thought to begin just after or perhaps within a short tandem repeat at the 5′ distal portion of the EPRV enhancer (Kanno et al., 2008). Coilin, which binds preferentially to small noncoding RNAs containing stem-loop structures (Machyna et al., 2014), could conceivably have an affinity for potential stem-loop structures in the 5′-UTR of GFP RNA if one or more repeat monomer sequences are present in the pre-mRNA (Figure S7). Given the proposed function of coilin in snRNP biogenesis and splicing, it is possible that some unspecified feature of the alternative splicing pattern of GFP pre-mRNA makes it a prime target of coilin activity. Alternative splicing was not deliberately engineered into the GFP transgene construct but only became apparent when trying to determine the transcription start site of the GFP reporter gene (Kanno et al., 2008). The CaMV 35S transcript exhibits a flexible and complex alternative splicing pattern (Bouton et al., 2015), and our findings suggest that this may be common feature of pararetroviral transcripts. However, despite possible connections among coilin, viruses, splicing, and RNAs with secondary structures, the reasons for the sensitivity of the GFP reporter gene to coilin function remain unknown. The well-defined nature of the GFP reporter gene system should enable dissection of the sequence features critical for coilin recognition and function.
Coilin has been described as a “global sensor that responds to environmental signals” (Hebert 2013). Our results are compatible with this notion in so far as stress- and senescence-related genes appear to be common among the differentially expressed and more efficiently spliced genes in coilin mutants. However, the coilin mutants did not consistently display altered responses to the stress treatments tested (salt, chilling, and RNA virus infection). Perhaps subtle effects occurred that were not readily apparent in our experiments. More detailed analyses of stress responses and additional numbers of stress tests on the coilin mutants may reveal clearer patterns of differential stress tolerance compared with wild-type plants. Coilin knockdown in N. benthamiana plants was reported to variably alter interactions with different viruses, either enhancing or reducing pathogenicity (Shaw et al., 2014). However, the four Arabidopsis coilin mutants we tested responded similarly to wild-type plants when infected with TuMV or CMV, which are both RNA viruses. TuMV is a potyvirus, a type that showed impaired infection after coilin knockdown in N. benthamiana (Shaw et al., 2014). The differing results obtained in the two studies may reflect varying susceptibility to viral infection following coilin depletion in the two different plant species. Future work can extend these investigations by testing the responses of Arabidopsis coilin mutants to a larger selection of viruses.
Of the coilin mutations recovered in our screen, two resulted in amino acid substitutions in the most highly conserved regions of the protein: the N-terminal self-association domain (E76K) and the C-terminal Tudor-like domain (P439L). The self-association domain, which comprises around 90 amino acids, is required for coilin to self-interact and to be targeted to CBs (Hebert and Matera 2000). Unlike typical Tudor domains, the Tudor-like domain in coilin does not appear to bind directly to methylated amino acids (Shanbhag et al., 2010), but it has been shown to interact with Sm proteins, which make up the stable core of the snRNP complex (Xu et al., 2005), and survival of motor neuron (SMN), a CB protein essential for the assembly of snRNPs (Hebert et al., 2001). The positions of the loss-of-function mutations we recovered reinforce the importance of the two conserved domains for coilin function and suggest important amino acids on which to focus in future studies.
Our results suggest a broad view of coilin activity that involves relatively subtle influences on transcription and splicing by mechanisms that remain to be determined. Coilin may fine tune gene expression at multiple levels by means of its ability to bind nucleic acids or to recruit other proteins to the transcriptional or spliceosomal machinery. In the wild-type T line, coilin acts to attenuate expression of the GFP reporter gene, which clearly has the potential to be expressed at a higher level than that normally sustained. This suggests a rheostat-like function for coilin, but how this may work is unclear. Moreover, given the complete deficit of normal coilin protein and dispersal of CBs in our mutants, we are unable to assess whether increased expression of the GFP reporter gene reflects CB-dependent or nucleoplasmic functions of coilin.
Further insights into the molecular mechanisms of coilin function in plants can be gained by investigating coilin-associated genes and coilin-interacting proteins. In animal cells, coilin has been shown to associate with histone, snoRNA, and snRNA genes (Machyna et al., 2014, 2015) and to interact with the aforementioned Sm and SMN proteins, which have orthologs in plants (Reddy et al., 2013). In plants, siRNA and miRNA biogenesis occurs in CBs (Pontes and Pikaard 2008). Thus, sequencing of size-selected RNAs from Arabidopsis coilin mutants may reveal unique functions of this protein in plant small RNA metabolism. A recent global mapping of protein and RNA interaction partners of coilin in animal cells did not reveal surprisingly novel proteins or RNAs, suggesting that interacting partners for coilin are known and that the molecular function should soon be clarified (Machyna et al., 2015). The GFP reporter gene system, which provides a convenient readout for coilin activity that does not rely on monitoring CB integrity, should be useful for probing further the functions of this elusive protein in a plant system.
Acknowledgments
We thank Fang-Fang Chen for assistance with the virus infection experiments. Financial support for this research was provided by the Institute of Plant and Microbial Biology, Academia Sinica, and grants from the Taiwan Ministry of Science and Technology to M.M. and A.M. (MOST 103-2311-B001-004-MY3 and MOST 104-2311-B-001-037).
Footnotes
Communicating editor: S. M. Springer
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.190751/-/DC1.
Literature Cited
- Andrade L. E., Chan E. K., Raska I., Peebles C. L., Roos G., et al. , 1991. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 173: 1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin R. S., Vidaurre D., Stamatiou G., Breit R., Provart N. J., et al. , 2011. Next generation mapping of Arabidopsis genes. Plant J. 67: 715–725. [DOI] [PubMed] [Google Scholar]
- Austin R. S., Chatfield S. P., Desveaux D., Guttman D. S., 2014. Next generation mapping of genetic mutations using bulk population sequencing. Methods Mol. Biol. 1062: 301–315. [DOI] [PubMed] [Google Scholar]
- Bellini M., 2000. Coilin, more than a marker of the Cajal (coiled) body. BioEssays 22: 861–867. [DOI] [PubMed] [Google Scholar]
- Benfey P. N., Ren L., Chua N. H., 1989. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 8: 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudonck K., Dolan L., Shaw P. J., 1998. Coiled body numbers in the Arabidopsis root epidermis are regulated by cell type, developmental stage and cell cycle parameters. J. Cell Sci. 111: 3687–3694. [DOI] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ. (1999) The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol. Biol. Cell 10(7): 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton C., Geldreich A., Ramel L., Ryabova L. A., Dimitrova M., et al. , 2015. Cauliflower mosaic virus transcriptome reveals a complex alternative splicing pattern. PLoS One 10: e0132665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E., Harrison E., McHattie S., Hughes L., Hickman R., et al. , 2011. High resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 8738–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome H. J., Hebert M. D., 2012. In vitro RNase and nuclei acid binding activities implicate coilin in U snRNA processing. PLoS One 7: e36300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M., Lamond A. I., 2005. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 21: 105–131. [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collier S., Pendle A., Boudonck K., van Rij T., Dolan L., et al. , 2006. A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol. Biol. Cell 17: 2942–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enwerem I. I., Wu G., Yu Y. T., Hebert M. D., 2014. Cajal body proteins differentially affect the processing of box C/D scaRNPs. PLoS One 10: e0122348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J. L., Kanno T., Liang S. C., Matzke A. J., Matzke M., 2015. GFP loss-of-function mutations in Arabidopsis thaliana. G3 (Bethesda) 5: 1849–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor W., Mette M. F., Staginnus C., Matzke M. A., Matzke A. J., 2004. A distinct endogenous pararetrovirus family in Nicotiana tomentosiformis, a diploid progenitor of polyploid tobacco. Plant Physiol. 134: 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G. W., Somerville C. R., 1990. A mutation causing imidazolinone resistance maps to the Csr1 locus of Arabidopsis thaliana. Plant Physiol. 92: 1081–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P., 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–999. [DOI] [PubMed] [Google Scholar]
- Hebert M. D., 2010. Phosphorylation and the Cajal body: modification in search of function. Arch. Biochem. Biophys. 496: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert M. D., 2013. Signals controlling Cajal body assembly and function. Int. J. Biochem. Cell Biol. 45: 314–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert M. D., Matera A. G., 2000. Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 11: 4159–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert M. D., Szymczyk P. W., Shpargel K. B., Matera A. G., 2001. Colin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 15: 2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzel L., Neugebauer K. M., 2015. Quantification of co-transcriptional splicing from RNA-seq data. Methods 85: 36–43. [DOI] [PubMed] [Google Scholar]
- Ito H., Kim J. M., Matsunaga W., Saze H., Matsui A., et al. , 2016. A stress-activated transposon in Arabidopsis induces transgenerational abscisic acid insensitivity. Sci. Rep. 6: 23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James N. J., Howell G. J., Walker J. H., Blair G. E., 2010. The role of Cajal bodies in the expression of late phase adenovirus proteins. Virology 399: 299–311. [DOI] [PubMed] [Google Scholar]
- Kanno T., Bucher E., Daxinger L., Huettel B., Böhmdorfer G., et al. , 2008. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat. Genet. 40: 670–675. [DOI] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Schumaker K. S., Zhu J. K., 2006. EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323: 101–103. [DOI] [PubMed] [Google Scholar]
- Kung Y. J., Lin P. C., Yeh S. D., Hong S. F., Chua N. H., et al. , 2014. Genetic analyses of the FRNK motif function of turnip mosaic virus uncover multiple and potentially interactive pathways of cross-protection. Mol. Plant Microbe Interact. 27: 944–955. [DOI] [PubMed] [Google Scholar]
- Lam Y. W., Lyon C. E., Lamond A. I., 2002. Large-scale isolation of Cajal bodies from HeLa cells. Mol. Biol. Cell 13: 2461–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Li W., Lin W. D., Santi S., Schmidt W., 2013. Mapping gene activity of Arabidopsis root hairs. Genome Biol. 14: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K., 2014. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. L., Wu Z., Nizami Z., Deryusheva S., Rajendra T. K., et al. , 2009. Coiln is essential for Cajal body organization in Drosophila. Mol. Biol. Cell 20: 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M., Kehr S., Straube K., Kappei D., Buchholz F., et al. , 2014. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol. Cell 56: 389–399. [DOI] [PubMed] [Google Scholar]
- Machyna M., Neugebauer K. M., Staněk D., 2015. Coilin: the first 25 years. RNA Biol. 12: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov V., Rakitina D., Protopopova A., Yaminsky I., Arutiunian A., et al. , 2013. Plant coilin: structural characteristics and RNA binding properties. PLoS One 8: e53571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Koester T., Staiger D., 2015. Pre-mRNA splicing in plants: in vivo functions of RNA binding proteins implicated in the splicing process. Biomolecules 5: 1717–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q. W., Lin S. S., Reyes J. L., Chen K. C., Wu H. W., et al. , 2006. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Novotný I., Malinová A., Stejskalová E., Matějů D., Klimešová K., et al. , 2015. SART3-dependent accumulation of incomplete spliceosomal snRNPs in Cajal bodies. Cell Reports 10: 429–440. [DOI] [PubMed] [Google Scholar]
- Pontes O., Pikaard C. S., 2008. siRNA and miRNA processing: new functions for Cajal bodies. Curr. Opin. Genet. Dev. 18: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra T. K., Praveen K., Matera A. G., 2010. Genetic analysis of nuclear bodies: from nondeterministic chaos to deterministic order. Cold Spring Harb. Symp. Quant. Biol. 75: 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I., Andrade L. E., Ochs R. L., Chan E. K., Chang C. M., et al. , 1991. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 195: 27–37. [DOI] [PubMed] [Google Scholar]
- Reddy A. S., Marquez Y., Kalyna M., Barta A., 2013. Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A., 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Lee T. F., Liao W. W., Naumann U., Liao J. L., et al. , 2014. Distinct and concurrent pathways of Pol II- and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana. Plant J. 79: 127–138. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Kanno T., Liang S. C., Chen P. Y., Liao W. W., et al. , 2015. An Rtf2 domain-containing protein influences pre-mRNA splicing and is essential for embryonic development in Arabidopsis thaliana. Genetics 200: 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag R., Kurabi A., Kwan J. J., Donaldson L. W., 2010. Solution structure of the carboxy-terminal Tudor domain from human coilin. FEBS Lett. 584: 4351–4356. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Burge C. B., 1997. Classification of introns: U2-type or U12-type. Cell 91: 875–879. [DOI] [PubMed] [Google Scholar]
- Shaw J., Love A. J., Makarova S. S., Kalinina N. O., Harrison B. D., et al. , 2014. Coilin, the signature protein of Cajal bodies, differentially modulates the interactions of plants with viruses in widely different taxa. Nucleus 5: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M., Trowitzsch S., Weber G., Lührmann R., Oates A. C., et al. , 2010. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat. Struct. Mol. Biol. 17: 403–409. [DOI] [PubMed] [Google Scholar]
- Verslues P. E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J. K., 2006. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45: 523–539. [DOI] [PubMed] [Google Scholar]
- Walker M. P., Tian L., Matera A. G., 2009. Reduced viability, fertility and fecundity in mice lacking the Cajal body marker protein, coilin. PLoS One 4(7): e6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., et al. , 2007. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2(8): e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittom A. A., Xu H., Hebert M. D., 2008. Coilin levels and modifications influence artificial reporter splicing. Cell. Mol. Life Sci. 65: 1256–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Pillai R. S., Azzouz T. N., Shpargel K. B., Kambach C., et al. , 2005. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 114: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Seeds of the wild-type T line and the coilin mutants will be deposited at the Arabidopsis Biological Resource Center, and are currently available on request from Matzke’s laboratory. RNA-seq data are available from NCBI SRA under accession number SRP071829.