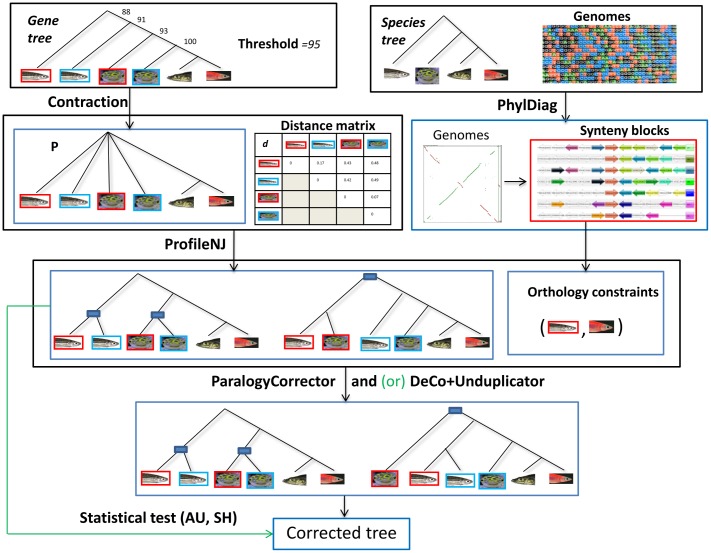
Fig 5. A general view on RefineTree when run on the Ensembl Compara gene families.
An example is given for a species tree S of four fish species, a gene family of six genes (a gene is represented by the picture of the species it belongs to, and two paralogs belonging to the same species are distinguished by a different frame color), a rooted gene tree G (although it can be non rooted in general) with branch support, and a given threshold for branch contraction. Data framed in black are the input and those framed in blue are the output of the correction algorithm labeling the edge linking the considered frames. Black arrows depict the use we make of RefineTree on the Ensembl gene trees. The green arrow and the green “or” are alternative uses avoiding one or both of the correction tools ParalogyCorrector and Unduplicator. Any framed set of data can be alternatively provided to the pipeline as input. For example, orthology constraints obtained from various sources can be directly provided as input to ParalogyCorrector. The method for inferring orthology constraints from synteny blocks is described in the text.