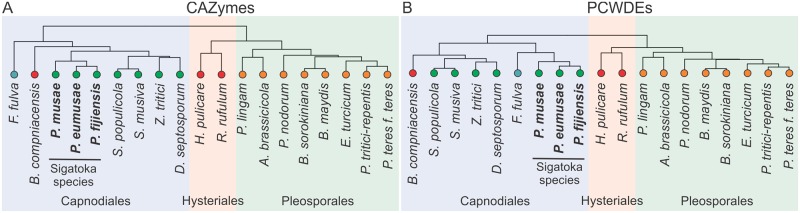
Fig 7. Hierarchical clustering of Pseudocercospora musae, Pseudocercospora eumusae, Pseudocercospora fijiensis, and 16 other representative Dothideomycete fungi with different nutritional lifestyles, based on copy number changes in carbohydrate-active enzyme (CAZyme) families or the subset of plant cell wall degrading enzymes (PCWDEs).
The selected 16 representative Dothideomycete species that are included in the analysis fall into three major orders: Capnodiales (red), Hysteriales (blue), and Pleosporales (green). The nutritional lifestyle of each species is indicated by a colored dot next to each species name: biotrophs (blue), hemi-biotrophs (green), necrotrophs (yellow), saprophytes (red). (A) Hierarchical clustering of the species based on their total CAzyme distribution profile (i.e. the number of genes assigned to each CAZyme family) (B) Hierarchical clustering of the species based on their distribution profile for CAZyme families related to plant cell wall degradation. In both cases, clustering supported a swapped topology in which P. eumusae is clustered together with P. fijiensis, suggesting that these two species share a more similar pattern of gene family expansions and contractions in CAZymes and PCWDEs in particular.