HOT TOPIC
Mutations in leucine-rich repeat kinase (LRRK2) are one of the most common known genetic causes of neurodegeneration and responsible for many cases of late-onset Parkinson disease (PD). Emerging data from Multiethnic Exome Sequencing efforts (http://exac.broadinstitute.org) suggest that the common pathogenic LRRK2 mutation, G2019S, might be much more common than previously thought. Several genome-wide association studies and meta-analyses implicate LRRK2 in susceptibility to sporadic PD. The encoded LRRK2 protein is a complex enzyme considered to be part of the druggable proteome, and the G2019S LRRK2 mutation upregulates protein kinase activity that catalyzes the transfer of phosphate from ATP to a serine or threonine in a protein substrate1. The mechanisms of LRRK2 kinase activity that might contribute to neurodegeneration have not been fully elucidated, and a major bottleneck towards this understanding has been the lack of knowledge of trans LRRK2 kinase substrates. Bone fide LRRK2 kinase substrates can be defined as protein substrates that depend on LRRK2 kinase activity for their phosphorylation status in cells and tissue. In a recent publication, Steger et al present convincing datasets that show several phosphopeptides in Rab proteins can be regulated by LRRK2 kinase activity2.
One critical observation by Steger et al. is that all of the pathogenic LRRK2 mutations, whether in the LRRK2 Rab-like GTPase domain (e.g, R1441C), the adjacent c-terminal of ROC domain (Y1699C), or the kinase domain (e.g., G2019S), all increase threonine phosphorylation of the Rab protein substrates Rab10 and Rab8 in cells. The notion that LRRK2 pathogenic mutations increase kinase activity in cells is also supported by recent observations related to cis-LRRK2 phosphorylation (i.e., LRRK2 autophosphorylation). Autophosphorylation of S1292 in LRRK2 is, like Rab phosphorylation, increased by all pathogenic LRRK2 mutations3, 4. The identification of bone fide trans and cis LRRK2 kinase targets allows, for the first time, the analysis of cells and tissue most vulnerable in PD in both model systems and biospecimens from patients.
In contrast to cell based assays, Steger et al. report that in vitro kinase assays with the Rab substrates do not reveal a difference in threonine phosphorylation between wild-type LRRK2 and the pathogenic R1441C or Y1699C LRRK2. Along the same lines, serine phosphorylation in switch II of another subset of Rabs, including S105 of Rab12, that are not direct substrates of LRRK2 in vitro, depend on LRRK2 activity and are increased by all LRRK2 mutations tested (Figure 1). An important difference therefore emerges between in vitro LRRK2 kinase assays and the activity of LRRK2 in cells. The reason for the discrepancy between in vivo and in vitro results is not clear at this time, but would be important to pursue. One possibility is that the two enzymatic activities of LRRK2, namely GTPase and kinase activity, co-operate in cells to control the overall phosphorylation of Rab proteins. Alternatively, there may be modifier proteins or co-factors that interact with LRRK2 and influence cellular function that are currently missing in the reductionist in vitro assays widely employed. Both of these hypotheses are testable, and the scope of therapeutic indications for LRRK2-targeted therapeutics may widen commensurate with the newly attributed range of LRRK2 activities.
Figure 1.
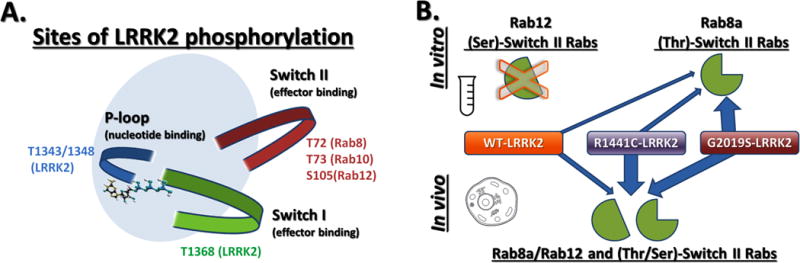
A) Threonine or serine residues in Rab receptacles that depend on LRRK2 kinase activity for their phosphorylated state in cells. B) Rabs with serine in switch II residues may not serve as efficient LRRK2 kinase substrates for LRRK2 in vitro but are regulated by LRRK2 in cells. Rabs with threonine residues in switch II may serve as direct LRRK2 kinase substrates both in vitro and in cells. Blue arrows indicate relative strength of LRRK2-mediate phosphorylation of the switch II receptacles.
Another major conclusion of Steger et al. is that LRRK2 protein may regulate Rab GTPases that are important in many signaling cascades through phosphorylation. The Ras superfamily of GTPases that includes the Rab family encodes conserved stretches of amino acids that create receptacles for effector proteins called the switch regions, composed of switch I, switch II, and the P-loop. The LRRK2 protein encodes all three of these effector binding sites and aligns very well to the Rab family. LRRK2 can add phosphosphates to its own switch I and P-loop receptacles through autophosphorylation and this process enhances GTPase activity5. With the trans LRRK2 Rab substrates Rab8 and Rab10, Steger et al. found that switch II phosphorylation results in diminished binding to both guanosine nucleotide dissociation inhibitor (GDI) that prevents nucleotide exchange as well as diminished binding to guanosine exchange factors (GEF) that enhances nucleotide exchange. There are few published studies attempting to determine the functional effects of phosphorylation of any GTPase. A priori prediction of the effects of phosphorylation on a GTPase receptacle domain are impossible because the functional outcome would depend on the constituency and availability of the local interacting effector proteins as well as intrinsic properties of the GTPase. In some GTPases with extremely high nucleotide off rates like LRRK2, GDIs but not GEFs would be critically important for GTPase activation, whereas in GTPases with very slow nucleotide off rates and high affinities, GEFs would be physiologically relevant. Meaningful effects on LRRK2 regulation of Rab GTPases will need to be established under a variety of conditions for every GTPase to help understand whether a particular Rab phosphorylation matters for pathways that underlie neurodegeneration.
Finally, Steger et al present a novel conceptualization and implementation of an unbiased high-powered approach to identify novel kinase substrates and validation of this process by the identification of at least three bone fide novel LRRK2 kinase substrates. In future studies, it would be important to empirically determine the false-discovery rate associated with these phosphoproteomic datasets so that true LRRK2 kinase substrates can be distinguished from phosphopeptides identified due to type I errors. The choice of mouse embryonic fibroblasts (MEF) by the authors as a vehicle cell model to identify LRRK2 kinase substrates may limit the number of candidate substrates that are identified. To date, LRRK2 activity in fibroblasts has not been shown to have physiological or phenotypic effects, in contrast to numerous functional phenotypes associated with LRRK2 expression in neurons and macrophages. In addition, LRRK2 cis autophosphorylation sites that have been validated using phospho-specific antibodies (e.g., pS1292) as well as appear in the phospho-proteome database generated by unbiased mass spectrometry screens (e.g., pT1357, http://www.phosphosite.org) were not detected by the authors in their approaches, suggesting either very low LRRK2 expression in these cells or very low LRRK2 kinase activity. Whether the approach by Steger et al. represents a generalizable method for future LRRK2 kinase substrate discovery at the moment is not clear but suggests that additional screens in other tissues might yield further insights.
The identification of LRRK2 Rab kinase substrates may be a very important advance in understanding the role of LRRK2 in PD. In pursing LRRK2 as a therapeutic target in PD, knowledge of trans kinase substrates may be important in both patient selection for LRRK2 kinase inhibitors as well as pharmacodynamic assays. With the combination of high quality LRRK2 small molecule inhibitor tools, LRRK2 monoclonal antibodies and rodents models, and now LRRK2 kinase substrates, novel therapeutics that target LRRK2 are better poised for success in clinical trials for neuroprotection in PD.
Acknowledgments
This research was supported by NIH-National Institute of Neurologic Disorders and Stroke (U01 NS097028 and R01 NS064934), and the Intramural Research Program of the NIH, National Institute on Aging.
References
- 1.West AB, Moore DJ, Biskup S, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(46):16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steger M, Tonelli F, Ito G, et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. Elife. 2016;5 doi: 10.7554/eLife.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reynolds A, Doggett EA, Riddle SM, Lebakken CS, Nichols RJ. LRRK2 kinase activity and biology are not uniformly predicted by its autophosphorylation and cellular phosphorylation site status. Front Mol Neurosci. 2014;7:54. doi: 10.3389/fnmol.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheng Z, Zhang S, Bustos D, et al. Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci Transl Med. 2012;4(164):164ra161. doi: 10.1126/scitranslmed.3004485. [DOI] [PubMed] [Google Scholar]
- 5.Liu Z, Mobley JA, DeLucas LJ, Kahn RA, West AB. LRRK2 autophosphorylation enhances its GTPase activity. FASEB J. 2016;30(1):336–347. doi: 10.1096/fj.15-277095. [DOI] [PMC free article] [PubMed] [Google Scholar]


