Abstract
Background
In C. elegans, germline development and early embryogenesis rely on post-transcriptional regulation of maternally transcribed mRNAs. In many cases, the 3′UTR is sufficient to govern the expression patterns of these transcripts. Several RNA-binding proteins are required to regulate maternal mRNAs through the 3′UTR. Despite intensive efforts to map RNA-binding protein-mRNA interactions in vivo, the biological impact of most binding events remains unknown. Reporter studies using single copy integrated transgenes are essential to evaluate the functional consequences of interactions between RNA-binding proteins and their associated mRNAs.
Results
In this report, we present an efficient method of generating reporter strains with improved throughput by using a library variant of MosSCI transgenesis. Furthermore, using RNA interference, we identify the suite of RBPs that control the expression pattern of five different maternal mRNAs.
Conclusions
The results provide a generalizable and efficient strategy to assess the functional relevance of protein-RNA interactions in vivo, and reveal new regulatory connections between key RNA-binding proteins and their maternal mRNA targets.
Keywords: C. elegans, MosSCI, RNA-binding proteins, 3′UTR, post-transcriptional regulation, OMA-1, DAZ-1, atg-4.2, cul-4, ets-4, set-6, mex-3, set-2
Introduction
Spatial and temporal regulation of gene expression is crucial to the differentiation of tissues and organs. Cell fate specification, axis formation, cleavage and cell division rely upon regulated expression of important gene products at the right place at the right time. Changes in gene expression patterns can be regulated at the level of transcription, splicing, nuclear export, localization, translation or stability of mRNAs and proteins (Jacobson and Peltz, 1996; Lasko, 2003; Melton, 1987; Moore, 2005; Thompson et al., 2007; Wickens et al., 2000).
In early embryogenesis, there is little or no active transcription in the zygotic nucleus (Batchelder et al., 1999; Leatherman and Jongens, 2003; Newport and Kirschner, 1982; Tadros and Lipshitz, 2009); hence post-transcriptional regulation of maternal mRNAs by RNA-binding proteins plays a critical role in several systems (Colegrove-Otero et al., 2005; Farley and Ryder, 2008; Spirin, 1966). There are many examples that illustrate this point. In Drosophila melanogaster, repression of hunchback mRNA by Nanos and Pumilio, and repression of caudal mRNA by Bicoid, is required for anterior-posterior axis formation (Dean et al., 2002). The spatial pattern of translation and repression is mediated by elements present in the 3′ untranslated regions (3′UTRs) of target transcripts. In Xenopus laevis, cyclinB1 mRNA must be kept translationally repressed in immature oocytes (Barkoff et al., 1998; de Moor, 1999). The RNA binding protein CPEB acts through the 3′UTR binding elements to mediate this repression until oocyte maturation. In Caenorhabditis elegans, sexual fate of gametogenesis relies on post-transcriptional regulation of fem-3 mRNA. Repression of fem-3 by FBF-1 and FBF-2 RNA-binding proteins is required for the switch from spermatogenesis to oogenesis to occur after the L4 to adult molt in the hermaphroditic worm (Zhang et al., 1997). These examples highlight the key conserved role of post-transcriptional regulation of maternal transcripts during metazoan development.
C. elegans germline development is an excellent model system to study post-transcriptional regulation. Development can be monitored in real time by light microscopy because the animal is transparent. Gene regulation can also be visualized in live nematodes using fluorescent reporter proteins (Chalfie et al., 1994), During oogenesis, transcription ceases when the oocytes enter prophase arrest. Transcription is not activated until the four-cell stage embryo, and then only in the somatic blastomeres (Blackwell, 2006; Seydoux and Fire, 1994; Seydoux et al., 1996; Walker et al., 2007). Therefore, maternal mRNAs transcribed by the germ cell nuclei must be controlled in the germline, in oocytes, and early embryos to regulate complex events of meiosis, oogenesis and early cell divisions in the embryo (Ahringer, 1997; Farley and Ryder, 2008; Seydoux, 1996; Stitzel and Seydoux, 2007). Specific RNA-binding proteins (RBPs) regulate the timing and localization of maternal mRNA translation and this regulation is conferred through specific elements in the 3′UTR of maternal mRNAs (de Moor et al., 2005; Evans and Hunter, 2005).
The importance of the 3′UTR to regulation of maternal transcripts in the C. elegans germline is well established (Ahringer et al., 1992; Marin and Evans, 2003; Merritt et al., 2008; Mootz et al., 2004; Wickens et al., 2002; Zhang et al., 1997). Seydoux and coworkers showed that the 3′UTR is sufficient to govern the expression patterns of most maternal transcripts in the germline and early embryos (Merritt et al., 2008). Reporter transgenes expressing GFP under the control of a pan-germline promoter and a gene-specific 3′UTR recapitulated the expression pattern of the endogenous protein in 24 out of 30 of transgenes. In contrast, patterned expression was not observed in the reporter strains containing a gene-specific promoter and an unregulated 3′UTR.
There are a number of RNA-binding proteins required for regulation of maternal mRNAs in the germline and early embryos. Examples include GLD-1, PUF-5/6/7, FBF-1/2, POS-1, OMA-1, MEX-3, MEX-5/6 (Detwiler et al., 2001; Farley and Ryder, 2012; Francis et al., 1995; Huang et al., 2002; Jones et al., 1996; Kaymak and Ryder, 2013; Lublin and Evans, 2007; Marin and Evans, 2003; Pagano et al., 2009; Ryder et al., 2004; Schubert et al., 2000; Shimada et al., 2002; Spike et al., 2014b; Tabara et al., 1999; Wickens et al., 2002; Zhang et al., 1997). Published studies have identified candidate regulatory targets and/or identified RNA sequence motifs recognized by each of these proteins (Bernstein et al., 2005; Farley and Ryder, 2012; Farley et al., 2008; Kaymak and Ryder, 2013; Pagano et al., 2007; Pagano et al., 2009; Ryder et al., 2004; Spike et al., 2014b). For example, POS-1 and GLD-1 regulate the expression of glp-1 mRNA in the posterior of the early embryo through association with motifs in the 3′UTR (Farley and Ryder, 2012). Similarly, MEX-3 regulates nos-2 translation in somatic cells of the early embryo through direct association with motifs present in the 3′UTR (Pagano et al., 2009). FBF regulates expression of cki-2, fem-3, and gld-1 translation through direct 3′UTR interactions as well (Kalchhauser et al., 2011; Wright et al., 2010). However, the complete network of RNA interactions has not been established for most of these proteins, nor is it clear whether binding leads to regulation in all cases. Transgenic reporter studies are required to evaluate the biological consequence of binding.
There are a few different methods of generating transgenic C. elegans lines. The first requires introduction of DNA through microinjection into the germline (Mello and Fire, 1995; Mello et al., 1991). DNA introduced in this manner forms an extrachromosomal array that is passed to the progeny of the injected animal. This method does not generate a stably inheritable line, unless genomic integration is induced through DNA damage caused by ionizing radiation or UV exposure. A disadvantage of this method is germline silencing of transgenes due to the presence of repetitive copies of the transgene in the array (Kelly et al., 1997; Mello and Fire, 1995; Mello et al., 1991). A more recent method is biolistic transformation (Praitis et al., 2001). This results in integration of the transgene, hence stable inheritance, but the integration site is approximately random, and often there are multiple integration events, making it difficult to compare reporter expressions between strains. The most recent advancement, termed Mos1-mediated single copy integration (MosSCI) was developed by Jorgensen and colleagues (Frøkjaer-Jensen et al., 2008; Frøkjaer-Jensen et al., 2012). In this method, DNA is microinjected into the germline, and site specific integration is induced through site specific DNA double strand break induction followed by homologous recombination. The major advantage of this method is that a single copy integration is generated at a predetermined chromosomal location. This method has been widely adopted to generate transgenic reporter strains where comparison of reporter expression patterns under different conditions is needed. Despite these improvements, MosSCI remains a time consuming approach that requires microinjection by a skilled microscopist.
In this study, we adapted the MosSCI method to simultaneously inject a library of transgenes into the gonad. We show that integration selects individual reporters from the injected pool, yielding numerous single copy integrants of different reporters. This approach has increased the efficiency of obtaining transgenic lines through limiting the number of injections necessary. We used this approach to generate twenty-one transgenic lines, including eleven unique 3′UTR reporter strains. We then performed a candidate RNAi screen using a subset of these strains to identify RBPs that control the pattern of expression. The results provide an enhanced method to rapidly generate transgenic nematode reporter strains, and identify new regulatory connections between maternal RBPs and maternal mRNAs.
Results
Library MosSCI
Motif analysis predicts the presence of numerous RBP binding sites in 3′UTRs (Farley et al., 2008; Keene, 2007; Pagano et al., 2007; Wright et al., 2010). However, functional studies show that only some binding sites are capable of conferring regulatory activity in cells and animals (Evans et al., 1994; Farley and Ryder, 2012; Kalchhauser et al., 2011; Marin and Evans, 2003; Pagano et al., 2009; Wright et al., 2010). We therefore sought a way to improve the throughput of functional studies using transgenic fluorescent reporters. The improved MosSCI protocol of Jorgensen lab was used as a starting point for our experiments (Frøkjaer-Jensen et al., 2012). We reasoned that the integration step of transgenesis might select individual transgenes from a library of reporter plasmids co-injected into worms. If so, then each injection could potentially create multiple transgenic progeny, each bearing a different integrated reporter transgene. To test this hypothesis, we cloned sixteen different 3′UTRs fused to a pan-germline promoter (mex-5) and a destabilized GFP-histone H2B fusion (GFP∷H2B∷PEST domain) (Farley and Ryder, 2012; Frand et al., 2005). The identities of the 3′UTRs used in transgenic reporter constructs are listed in Table 1.
Table 1.
List of the 3′UTRs in the reporters that were successfully integrated and their genotypes. The number of unique lines that were recovered for each reporter is also listed, as are the strain identifiers.
| 3′ UTR strain | Strain identifier | Genotype |
|---|---|---|
| EG6699 | ttTi5605 II; unc-119(ed3) III; oxEx1578 | |
| atg-4.2 | WRM10, WRM11 | sprSi10[Pmex-5∷MODC PEST:GFP:H2B∷ atg-4.2 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| cul-4 | WRM12, WRM13, WRM14, WRM15 | sprSi11[Pmex-5∷MODC PEST:GFP:H2B∷cul-4 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| cwn-1 | WRM16 | sprSi12[Pmex-5∷MODC PEST:GFP:H2B∷cwn-1 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| ets-4 | WRM17 | sprSi13[Pmex-5∷MODC PEST:GFP:H2B∷ets-4 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| hbl-1 | WRM18 | sprSi14[Pmex-5∷MODC PEST:GFP:H2B∷hbl-1 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| lin-26 | WRM19, WRM20, WRM21 | sprSi15[Pmex-5∷MODC PEST:GFP:H2B∷lin-26 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| mbk-2 | WRM22, WRM23 | sprSi16[Pmex-5∷MODC PEST:GFP:H2B∷mbk-2 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| mex-3 | WRM24, WRM25 | sprSi17[Pmex-5∷MODC PEST:GFP:H2B∷mex-3 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| set-2 | WRM26 | sprSi18[Pmex-5∷MODC PEST:GFP:H2B∷set-2 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| set-6 | WRM27, WRM28, WRM29 | sprSi19[Pmex-5∷MODC PEST:GFP:H2B∷set-6 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
| usp-14 | WRM30 | sprSi20[Pmex-5∷MODC PEST:GFP:H2B∷usp-14 3′UTR cb-unc-119(+)] II, unc-119(ed3) III |
Equal concentrations of each of the sixteen reporter constructs were mixed together into a single mixture. 25 ng/μl of this library mixture was injected into the C. elegans un-coordinated strain EG6699. Figure 1 shows a schematic of the injection method adapted from the MosSCI technique. After injecting the library of sixteen 3′UTR reporters constructs, the standard MosSCI protocol was followed (Frøkjaer-Jensen et al., 2012; Seidel et al., 2011). We then selected surviving worms and screened them and their progeny by PCR in order to identify which (if any) reporter construct had been integrated into the genome. We performed single worm PCR using a homozygous population of each candidate line. Figure 2 shows representative PCR data for the integrated lines obtained. Direct sequencing of the PCR products using a primer that anneals to the GFP reporter sequence led to unambiguous identification of which reporter construct had been integrated.
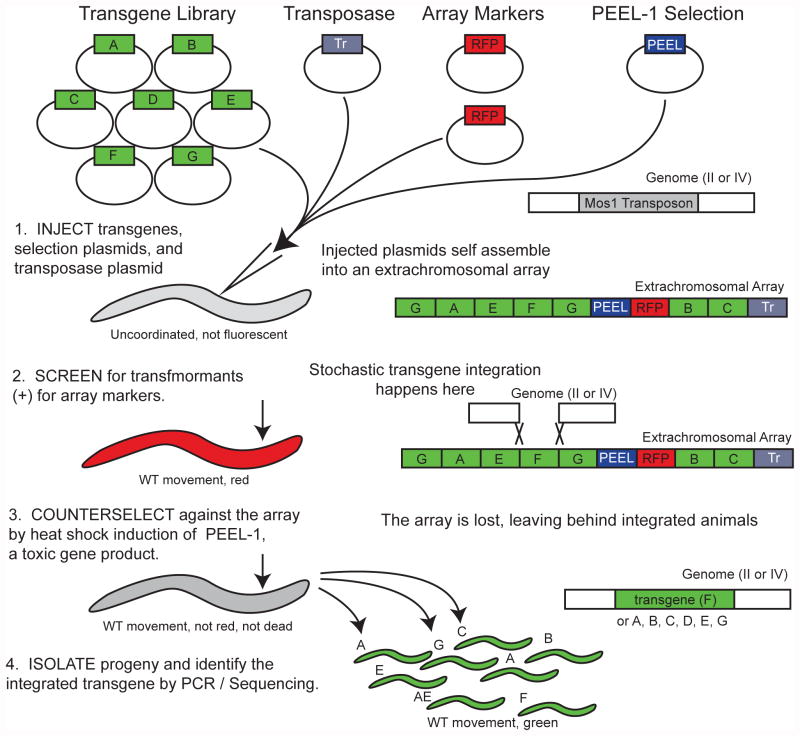
Figure 1.
Schematic of the library MosSCI method. A library of transgenes are mixed in equal amounts, and then microinjected into the germline of the parent strain. As with standard MosSCI, positive transformants are identified by unc-119 rescue, which restores wild-type movement, and red fluorescence in the pharynx and body wall muscle. Integrants are recovered by heat-shock induction of PEEL-1, a negative selection marker which kills all worms that have not lost extrachromosomal array formed from the injected plasmid mixture.
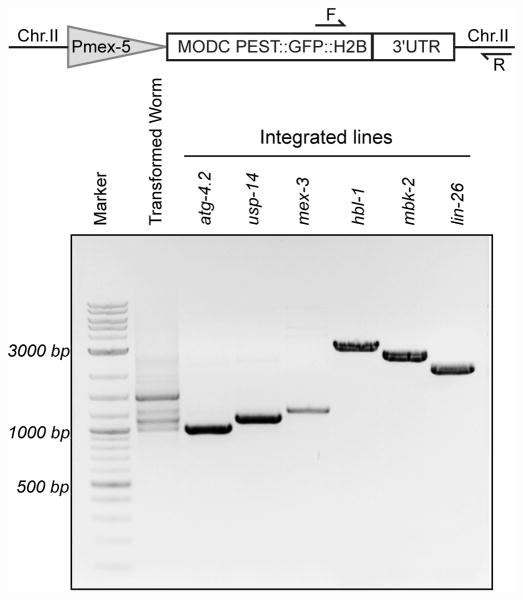
Figure 2.
Design of a single-worm based primer system to detect and characterize integrated lines. The first lane shows amplification from a transformed worm population, prior to counter-selection with PEEL-1, which harbors multiple reporters in an extrachromosomal array. The remaining lanes show the PCR amplified products of transgenes that were integrated. We determined which UTR reporter was integrated by sequencing the PCR products. The transgene schematic is not drawn to scale.
Fifteen transgenic lines were obtained from 217 injections. In this initial set of strains, we noticed that some constructs were integrated in more than one recovered line. Four of the integrated lines contained an integration of Pmex-5∷GFP∷H2B∷PEST∷cul-4 3′UTR. Three of the integrated lines contained Pmex-5∷GFP∷H2B∷PEST∷lin-26 3′UTR. Two of the integrated lines contained Pmex-5∷GFP∷H2B∷PEST∷mbk-2 3′UTR. Two more of the integrated lines contained Pmex-5∷GFP∷H2B∷PEST∷mex-3 3′UTR. Additional two of the integrated lines contained Pmex-5∷GFP∷H2B∷PEST∷atg-4.2 3′UTR. Of the fifteen recovered lines, we obtained seven unique reporters. To prevent recovering multiple copies of the same strain, we then reduced the size of the library to include just the remaining nine constructs from our library of sixteen. In additional 52 injections, we obtained six additional independent lines, resulting in four additional unique reporter strains. In total, we were able to generate eleven unique transgenic strains in 269 injections. These are listed in Table 1.
The rate of successful injections giving wild-type moving transformants was higher than the rate of integration steps. In the total of 269 attempted injections, 93 gave rise to wild-type moving transformant progeny. Of these, 21 contained a single copy of a transgenic construct. As such, we estimate our successful injection rate to be 35%, the integration rate to be approximately 23%, and the unique strain recovery rate (per successful injection for our relatively small library of sixteen reporters) to be ∼11%. We note that the overall transgenic recovery rate per injection is similar to previously reported values, suggesting that multiplexing reporters does not greatly impede integration. The original MosSCI protocol reported 8% integration rate, while a subsequent improved protocol reported a 54% integration rate (Frøkjaer-Jensen et al., 2008; Frøkjaer-Jensen et al., 2012).
While overall success is still limited by injection rate (governed by the ability of the injector), recovery of unique reporters was simplified due to the reduction in the overall number of plates that must be handled, and the time spent injecting animals. Fewer worms must be injected to achieve the total number of transgenic strains. For example in traditional MosSCI transgenesis, if 50 worms are injected per reporter transgene, 750 injections would be performed in order to make fifteen transgenic strains. Each successfully injected worm would need to be transferred to a single plate, and then monitored for integration following starvation. Our approach required just 269 injections to make fifteen reporter transgenes (eleven unique), and the concomitant decrease in the number of plates to be monitored, yielding approximately 50–66% reduction in injection and handling effort.
Expression patterns of GFP in the integrants
Having established new lines, we then used direct fluorescence imaging of the germline to determine the expression patterns of the transgenic reporter strains. Three out of the twenty-one lines we generated did not show GFP fluorescence, presumably due to germline transgene silencing (Kelly et al., 1997). The Mello lab showed that some transgenic constructs may be more prone to epigenetic silencing mechanisms in the germline than others whereby competition between two Argonautes, CSR-1 and PRG-1 determines whether a transgene is silenced or not. CSR-1 acquired sequences are expressed whereas PRG-1 acquired sequences are repressed. (Seth et al., 2013). The eighteen remaining strains prepared by library MosSCI showed GFP expression in the germline and/or embryos. As expected, the pattern of expression varied with the identity of the 3′UTR. The expression patterns are presented in Figure 3 and quantified in Figure 4.
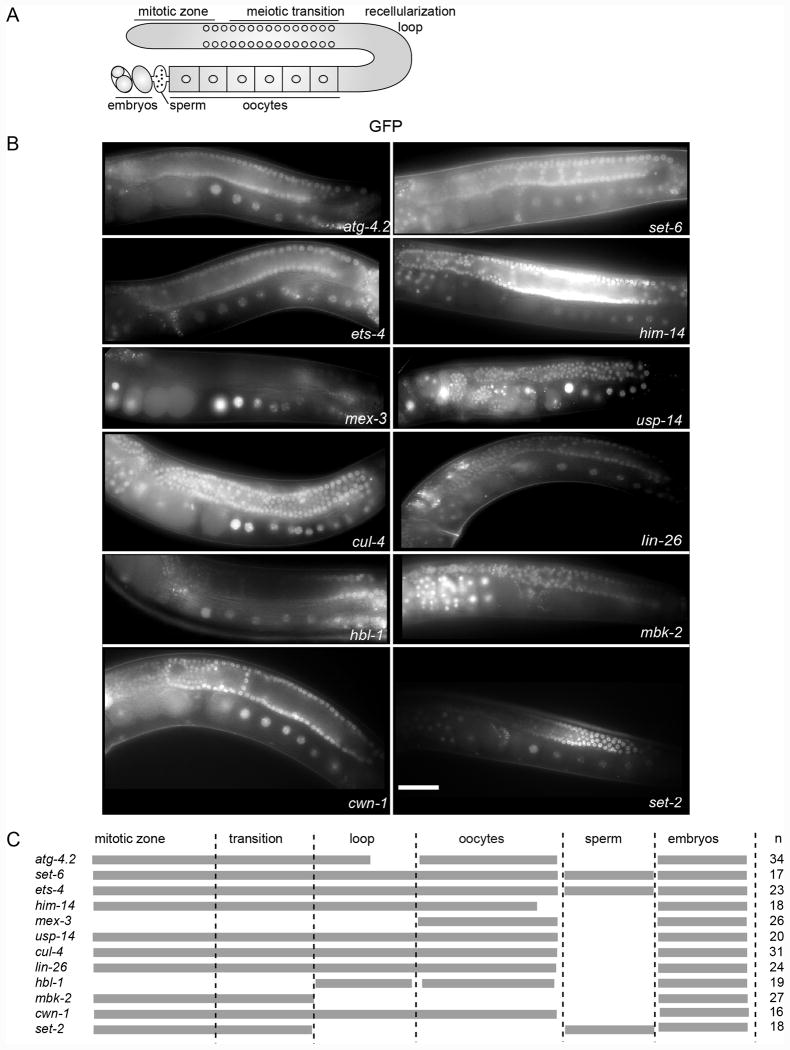
Figure 3.
GFP expression patterns of the 3′UTR reporter strains. Top: Schematic of the C. elegans germline. The syncytial region of nuclei in the distal arm of the gonad, the oocytes, sperm, and embryos in the uterus are shown. Middle: Representative images of single copy integrated reporter strains that express GFP under the control of different 3′UTRs. The white bar denotes 50 microns scale length. Bottom: A table summarizing the GFP expression patterns of the reporter strains in different parts of the germline and embryos. Gray bars denote expression. The number of animals imaged is indicated to the right.
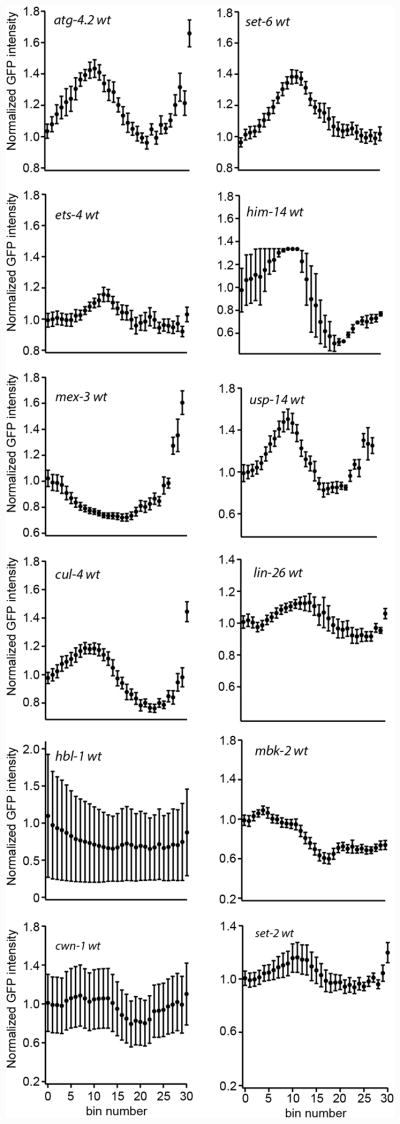
Figure 4.
Quantification of the mean GFP fluorescence intensity throughout the germline for strains presented in figure 3. The error bars denote the standard error of the mean intensity across bins defined by fractional length of the germline.
Pan germline expression
Some 3′UTR reporters showed pan-germline expression, including ets-4, usp-14, hbl-1, lin-26 and cwn-1. Reporter expression was observed in the distal region of the germline, in mitotic progenitor cells, as well as in the syncytial region, in the germline bend, in oocytes, and in embryos. We note that the hbl-1 reporter expression was faint in all regions of the germline. In four of the five reporters (usp-14, lin-26, hbl-1, and cwn-1), no expression was observed in sperm, consistent with the findings of Seydoux and co-workers that suggests sperm expression is governed via transcriptional regulation at the promoter, rather than post-transcriptionally through 3′UTR level (Merritt et al., 2008). In direct contrast, ets-4 reporter expression remained strong in sperm (n=12/23), suggesting that at least some 3′UTRs can direct retention of sperm specific expression.
The set-2 pattern
We also studied the pattern of a set-2 3′UTR that was integrated using standard reporter MosSCI, rather than the library approach presented here. set-2 3′UTR showed faint GFP expression in the distal end followed by an increased expression in the syncytial region, which then decreased around the recellularization region and oocytes. As with ets-4, the GFP expression remained strong in sperm (n=11/15), providing a second example of a 3′UTR that can direct expression of a reporter in male gametes.
Reduced oocyte expression
Other 3′UTR reporters, such as atg-4.2, cul-4, him-14 and set-6, and mbk-2 showed strong expression in the syncytial region of the gonad and in embryos but little or no expression in oocytes. atg-4.2 and cul-4 showed GFP expression in the distal mitotic zone followed by increased expression in the syncytial region, and decreased expression around the recellularization region and in early oocytes. Weak expression in oocytes appeared to increase as the oocytes neared the spermatheca. In contrast, set-6 and him-14 3′UTR reporters showed no increase in oocyte expression in maturing oocytes. Interestingly, set-6 reporter also showed GFP expression in sperm (n=10/17). Only the mbk-2 reporter showed a complete lack of expression in oocytes.
Oocyte- and embryo-specific expression
The mex-3 3′UTR reporter is unique in that it showed strong GFP expression in the oocytes, with expression peaking in the most mature oocytes. Little expression was observed in the distal germline or in the syncytial region. Expression was also observed in the anterior cells of early embryos, but not in the posterior. These results are consistent with the patterned expression of endogenous MEX-3 (Draper et al., 1996), and confirm the obsersvations made previously using a me-3 3′UTR reporter made by biolistic transformation (Merritt et al., 2008).
Out of the twelve 3′UTR reporter strains we studied, endogenous protein expression patterns are known for LIN-26, MEX-3, MBK-2 and SET-2. Antibody staining experiments showed that SET-2 and MEX-3 endogenous patterns match our reported patterns. MEX-3 is seen in the oocytes and anterior cells of two and four cell stage embryos matching our GFP reporter pattern (Bowerman et al., 1997; Draper et al., 1996). SET-2 is observed strongly in the mid-pachytene region of the germline but also in pharynx, neurons and intestines (Xu and Strome, 2001). We do not expect to observe somatic expression with our reporters, which include a germline specific promoter. No sperm expression was reported. For MBK-2, antibody staining was reported at the cortex of developing oocytes and in cytoplasm of embryos; however, we have not seen reporter expression in the oocytes of the mbk-2 3′UTR reporter strain (Stitzel et al., 2007). These differences could be due to transcriptional regulation by the endogenous promoter used, or due to post-translational regulation. LIN-26, on the other hand, is endogenously expressed in the somatic gonad and hypodermal cells of embryos and larvae of all stages; however, germline expression pattern was not reported (Labouesse et al., 1996). Endogenous protein expression patterns have not been published for ATG-4.2, CUL-4, HIM-14, ETS-4 SET-6, and USP-14.
Targeted RNAi screening of transgenic reporter strains
We wished to identify RNA-binding proteins that directly or indirectly control the expression pattern of the new 3′UTR reporter strains. We chose a subset of reporter strains that have distinct patterns of GFP expression to study further by RNAi knockdown studies. The strains we chose to investigate carry the 3′UTRs of the genes atg-4.2, cul-4, set-2, set-6, mex-3, or ets-4. In addition to their interesting patterns of expression, these 3′UTRs also contain binding motifs for RBPs with important roles in germline development and early embryogenesis. We wanted to identify which RNA-binding proteins contribute to the varying patterns of GFP expressions in the reporter strains. We looked for expression pattern changes under oma-1;oma-2 RNAi, daz-1 RNAi pos-1 RNAi, and control treatments. We chose to knockdown these transcripts because they encode germline expressed RNA-binding proteins that have an easy to score phenotype. oma-1;oma-2 RNAi, and daz-1 RNAi lead to sterility and pos-1 RNAi leads to embryonic lethality (Detwiler et al., 2001; Karashima et al., 2000; Tabara et al., 1999).
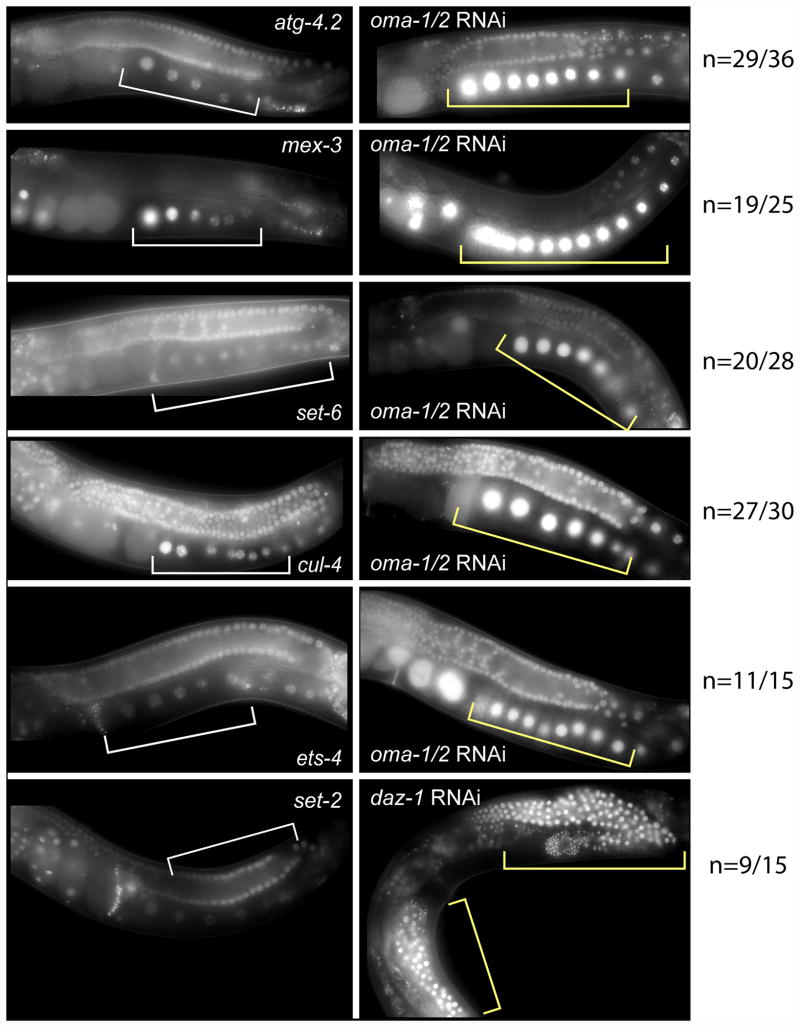
OMA-1 and OMA-2 are tandem zinc-finger RNA-binding proteins redundantly required for oocyte maturation. The phenotype of oma-1;oma-2 RNAi knockdown is more than 90% penetrant when performed by the feeding method. Knockdown of oma-1 and oma-2 by RNAi leads to oocytes with increased size, a greater number of oocytes in the gonad arm, and sterility (Detwiler et al., 2001; Lin, 2003). Knockdown in transgenic strains carrying the 3′UTRs of the genes atg-4.2, ets-4, cul-4, set-6, mex-3 led to a strong increase in the expression of GFP in oocytes (shown in Figure 5, quantified in Figure 6). In contrast, knockdown had no effect on the set-2 3′UTR reporter. The results suggest that OMA-1 and OMA-2 repress expression of atg-4.2, ets-4, cul-4, and set-6 in oocytes.
Figure 5.
RNAi reveals that OMA-1 and OMA-2 repress the expression of atg-4.2, mex-3, set-6, ets-4 and cul-4 reporters in oocytes, while DAZ-1 regulates repression of set-2 in the syncytial and loop regions of the germline. The left panel shows the expression pattern of the reporter fusion strain and the right panel shows the expression patterns of the worms cultured with bacteria that express double stranded RNA targeting the oma-1, oma-2, or daz-1 mRNAs. Changes in the pattern of reporter expression are annotated by comparing the white bars (left panel) to the yellow bars (right panel). The penetrance of the change in expression is indicated to the right of the images.
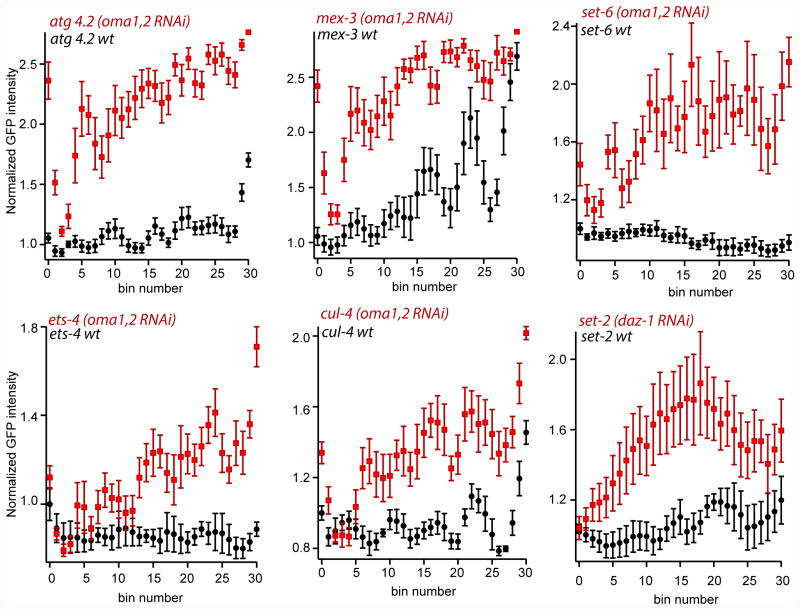
Figure 6.
Quantification of mean GFP fluorescence intensity for reporter strains treated with RNAi compared to empty vector control. The quantifications were performed as described in figure 4.
DAZ-1 is an RNA-binding protein required for oogenesis (Karashima et al., 2000; Otori et al., 2006). Knockdown of DAZ-1 results in absence of oocytes and sterility. The daz-1 RNAi-induced phenotype is 70-80% penetrant by the feeding method. Worms cultured under daz-1 RNAi conditions contain an abundance of non-cellularized nuclei around the germline bend, where oocytes normally form. This then leads to an absence of oocytes in the proximal region of the gonad arm. daz-1 RNAi was performed in strains carrying the atg-4.2 3′UTR, ets-4 3′UTR, cul-4 3′UTR, set-6 3′UTR, mex-3 3′UTR and set-2 3′UTR reporters. We observed a change only in the reporter strain containing the set-2 3′UTR. This strain does not express GFP around the loop region under wild-type conditions but when treated with daz-1 RNAi there was a strong increase in GFP expression in the recellularization/loop region (Fig. 5, Fig. 6). The results suggest that set-2, in contrast to atg-4.2, ets-4, cul-4, set-6, and mex-3, is regulated by DAZ-1, directly or indirectly.
POS-1 is another tandem zinc-finger RNA-binding protein that is required for the development of the posterior in the embryos. RNAi knockdown of this protein leads to embryonic lethality. The phenotype of pos-1 RNAi knockdown was about 80% penetrant. pos-1 RNAi knockdown did not show a change in the reporter expression in the germline and oocytes for any of the strains tested. In contrast, pos-1 knockdown has been previously shown to lead to expression of a glp-1 3′UTR reporter in all cells of an early embryo. The data suggest that POS-1 does not regulate many genes that harbor a putative POS-1 binding site, as has been previously suggested (Farley and Ryder, 2012).
Discussion
In this study, we have shown that the rate of generating transgenic strains can be improved using an adaptation to the MosSCI technique. Injecting a library of transgenic constructs reduced the total time consumed to make nineteen independent lines by three- to four-fold in our hands. This was achieved through stochastic integration of transgenic constructs for every successful injection. It is not yet clear if increasing the library size further will further improve the success rate. A larger sized library might introduce difficulties if one or more of the plasmids are toxic to the embryos. In such a case, elimination of the library construct one by one might be necessary to find which plasmid is causing the toxicity. Another potential disadvantage of the library MosSCI technique could be that, in theory, multiple transgenes can be integrated at the same time. To date, we have not yet observed either phenomonon.
The success rate of transgenesis is limited by the number of successful injections and by the extent of transgene integration. The rate of successful injections will vary between different injectors. The recent development of a microfluidic device to automate the injection procedure could help improve the number of successful injections (Gilleland et al., 2015). In this work, we used the direct insertion method of MosSCI. The extent of integration can be improved through the use of different promoters driving Mos1 transposase expression. For example, use of the eft-3 promoter has been shown to increase the rate of transformation presumably by increasing the extent of Mos1 transposon excision (Frøkjaer-Jensen et al., 2012). With this improvement, fewer injections may be sufficient to generate a number of strains after random integrations at the heat-shock step.
Obtaining transgenic strains at an increased rate will be advantageous in multiple ways. Library injection may be adapted to CRISPR-based approaches to make targeted mutations (DiCarlo et al., 2013; Friedland et al., 2013; Jinek et al., 2012; Kim et al., 2014; Marraffini and Sontheimer, 2010; Sorek et al., 2013; Wiedenheft et al., 2012). In an endogenous genomic locus of interest, a set of randomized insertions/deletions can be introduced through injection of a library of guide RNAs targeted for that locus. Using multiple CRISPR guides per injection can help ensure a mutation in the gene of interest, as has recently been shown in zebrafish (Gagnon et al., 2014).
In this study, we used library MosSCI to make 3′UTR reporters but this method could easily be adapted to make different promoter reporters or protein fusions to help define other aspects of regulatory biology, including transcription regulation and protein modification. A mutagenesis or deletion library analysis would help identify key cis-regulatory elements that control transcription regulation patterns critical to somatic differentiation in later stages of embryogenesis, after zygotic gene activation. Library MosSCI can also be used to rapidly generate mutants within a single UTR of interest and screen mutant strains to help map functional elements in a regulatory region of a UTR of interest. Another potential application of this technology could derive from systematically analyzing protein variants. Transgenic strains can be used to rescue a mutant phenotype by overexpressing a wild-type copy of the mutated gene. In such a case, injecting a library of overlapping fragments of the gene simultaneously could help identify the fragment that is minimally sufficient for rescue, if the overlapping fragments do not recombine.
Regulation by OMA-1/2
There are multiple ways OMA-1/2 could repress expression from the 3′UTR- reporters developed in our study. OMA-1/2 could be directly binding and repressing translation. Consistent with this hypothesis, each of the UTRs contain multiple UA(A/U) motifs recognized by OMA-1. OMA-1/2 could also be indirectly regulating transgene expression through antagonistic interactions with other key regulatory proteins. As shown in Spike et al., there are multiple proteins that associate with OMA-1/2 (Spike et al., 2014a; Spike et al., 2014b). The context of the OMA-1/2 RNP could affect regulatory activity. OMA-1 and OMA-2 repressed protein expression of most of the 3′UTR reporter transgenes we studied. This supports the hypothesis that OMA-1 might be a general repressor of translation during oocyte development and maturation (Kaymak and Ryder, 2013). The mRNAs that were regulated by OMA-1/2 encode proteins that influence a diverse array of biological phenomena. atg-4.2 encodes a homolog of human autophagic cysteine protease that does not have an obvious RNAi phenotype (Wu et al., 2012). ETS-4 is a transcription factor that regulates aging (Thyagarajan et al., 2010). CUL-4 is a cullin ubiquitin ligase that prevents re-replication of DNA (Zhong et al., 2003). set-6 is predicted to encode an H3K9 methyltransferase that regulates transcription (Andersen and Horvitz, 2007). mex-3 encodes a KH-domain RNA-binding protein that specifies the anterior of the embryo (Draper et al., 1996). As oocytes develop in the gonad arm, there is no autophagy, transcription, embryonic cell-fate determination or bulk DNA replication going on. This can be a reason why the mRNAs are kept in a silent state through OMA-1/2 acting as the major regulator or one of the intercommunicating regulators.
By contrast, oma-1, oma-2 RNAi did not repress the translation of the set-2 3′UTR reporter transgene. set-2 is a histone methyltransferase which can be involved in modifiying histones during chromatin remodeling which is required for the tight regulation of gene expression in sperm development (Simonet et al., 2007; Xiao et al., 2011). Intriguingly, DAZ-1 appears to regulate translation of the set-2 3′UTR. DAZ-1 is required for meiotic progression and formation of oocytes in the germline of C. elegans (Karashima et al., 2000; Otori et al., 2006). The RNA-binding specificity of DAZ-1 is not known, but its mammalian homolog DAZL (DAZ-like) binds stretches of polyU sequences with G or C bases distributed throughout ((G/CUn)n) (Venables et al., 2001). DAZ-1 represses set-2 3′UTR at the recellularization/loop region of the germline. set-2 is a methyltransferase that is required for proper germline development (Simonet et al., 2007; Xiao et al., 2011). It is not yet clear why this UTR is repressed by DAZ-1, but not OMA-1/2. More work is needed to understand why some transcripts are repressed by OMA-1/2 in oocytes, yet others are repressed by DAZ-1.
Sperm retention driven by the set-2, ets-4 and set-6 3′UTR
The Seydoux lab previously reported that promoters are necessary and sufficient for sperm expression for sperm-expressed reporter transgenes, while the 3′UTR sequence is dispensible for expression in sperm (Merritt et al., 2008). Here we show an exception to this finding where the 3′UTR of set-2, ets-4 and set-6 drives strong GFP expression in the sperm. Understanding how and why this 3′UTR enables expression in sperm may lead to new insights in sperm specific gene expression patterns. Moreover, we propose that incorporation of the set-2 or ets-4 3′UTR into a transgenic construct could provide a useful tool to enable studying the effect of driving expression of specific gene products in sperm.
Concluding remarks
The ability to generate transgenic strains in high yield will enable improved functional mapping of regulatory interaction networks between maternal mRNAs and RNA-binding proteins. Methods like CLIP, RIP-SEQ and PAR-CLIP identify interacting partners in vivo but may identify interactions that have no regulatory consequence. There are instances where an RNA-binding protein can play an active role in regulating a transcript through a binding site. In this case, the target site is necessary and sufficient for regulation. In other cases, the effect of an RNA-binding protein might be indirect or context dependent. In vivo studies with reporter strains carrying regulatory elements is necessary to distinguish between interactions of RNA-binding proteins that have a relevance to the regulation of an mRNA or not. As we have done in this work, the study of transgenic reporter strains carrying different C. elegans 3′UTRs can be done by RNAi screening. High-throughput RNAi screens could identify additional RNA-binding proteins that regulate these reporter transgenes. Once regulatory partners are identified, the necessity and sufficiency of target sites can be tested using library MosSCI to identify binding sites that are functionally important. Ultimately, the utility of large data sets that yield high resolution contact maps will be defined by their predictive power in functional studies. In order to keep pace, new technology to improve the output of functional studies in live animals is needed. Our work here demonstrates a simple strategy to improve the throughput of C. elegans single copy transgene strain production, a key first step towards this goal.
Experimental Procedures
Cloning of reporter constructs
The 3′UTR sequences were amplified from worm genomic DNA using UTR-specific primers flanked with atB2R and attB3 sequences for Gateway cloning. BP Clonase II was then used to clone the sequences into pDONRP2RP3. Multisite gateway cloning was then used to fuse this donor construct with pCM1.111 (construct carrying the mex-5 promoter) and pBMF2.7 (construct carrying MODC PEST:GFP:H2B). The PEST domain in the GFP construct helped reduce the half-life of GFP to prevent expansion of GFP expression to regions where it is not expressed due to its high stability (Farley and Ryder, 2012). LR Clonase II plus was used to integrate the fusion constructs into pCFJ150. Mutations of the glp-1 3′UTR reporter construct were introduced into the cloned pCFJ150 construct using Quickchange mutagenesis with Pfu Turbo.
C. elegans microinjection
MosSCI protocol (Frøkjaer-Jensen et al., 2008) was followed to generate the integrated lines with the adaptation described in this paper. A library of reporter constructs were injected at a concentration of 25ng/μl were injected with Peft-3∷mos-1 transposase (50 ng/μl), Pmyo-2∷mCherry (2.5 ng/μl), Pmyo-3∷mCherry and peel-1 (10 ng/μl) plasmids into the gonad of EG6699 un-coordinated strain at the young adult stage. Wild-type moving, surviving worms were screened by single-worm PCR using Pfu Ultra II to identify the integrated construct.
RNAi knockdown
We knocked down oma-1;oma-2, daz-1 and pos-1 using the RNAi feeding method (Kamath, 2003). Using TA cloning, we cloned the all oma-1 and oma-2 open reading frames (ORFs) into the vector L4440 and transformed the clones into HT115(DE3) cells. pos-1 and daz-1 RNAi feeding bacterial cells were obtained from the Ahringer library. At OD600 = 0.4, we induced the cells for four hours by adding 1 M isopropyl 1-thio-β-D-galactopyranoside (IPTG) at a final concentration of 0.4 mM. After induction, the induced RNAi cultures were concentrated 10- fold and added onto NGM plates containing 1mM IPTG and 100 μg/ml Ampicillin. Eggs obtained by bleaching adult worm strains were then plated onto RNAi plates and maintained at 25°C. The strains were imaged after 52 hours. HT115 strain bacteria transformed with the empty vector L4440 was used as our control plate.
Imaging of worm strains
Young adult worms were placed onto 2% agarose pads in 0.4 mM levamisole. DIC and GFP fluorescence images of gonad arms were taken using a 40× oil immersion objective (Zeiss Axioscope 2 plus microscope).
Quantifications of GFP intensities
For quantifications in Figure 4, GFP pixel intensities were determined by drawing a 7-pixel-wide segmented line passing through the nuclei starting from the distal end to the most mature oocyte. The average pixel intensity was determined across the line and divided into 30 equally sized bins and normalized as described previously (Farley and Ryder, 2012; Wright et al., 2010).
For quantifications in Figure 6, GFP pixel intensities were determined by drawing a 7-pixel-wide segmented line passing through the nuclei of daz-1 (RNAi) treated worms starting from the germline to the recellularization/loop region. For oma-1;oma-2 (RNAi) treated worms, GFP pixel intensities were determined by drawing a 20-pixel-wide segmented line passing through the nuclei of the oocytes. The same procedure was then followed to determine the average pixel intensity and compared to wild-type expression levels as described previously (Farley and Ryder, 2012; Kaymak and Ryder, 2013).
Acknowledgments
We thank Nick Rhind, Marian Walhout, and Craig Mello for helpful discussions and sharing of equipment. We also thank Takao Ishidate for assistance and training in C. elegans microinjections. Carina Clingman and Ruth Zearfoss provided helpful comments during the progress of this project. This work was supported in part by NIH Grants GM098643 and GM117237 to S.P.R.
Grant Sponsor: National Institutes of General Medical Sciences (NIGMS), Grant Numbers GM098643 and GM117237 (PI: Sean P. Ryder)
References
- Ahringer J. Maternal control of a zygotic patterning gene in Caenorhabditis elegans. Development. 1997;124:3865–3869. doi: 10.1242/dev.124.19.3865. [DOI] [PubMed] [Google Scholar]
- Ahringer J, Rosenquist TA, Lawson DN, Kimble J. The Caenorhabditis elegans sex determining gene fem-3 is regulated post-transcriptionally. EMBO J. 1992;11:2303–2310. doi: 10.1002/j.1460-2075.1992.tb05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- Barkoff A, Ballantyne S, Wickens M. Meiotic maturation in Xenopus requires polyadenylation of multiple mRNAs. EMBO J. 1998;17:3168–3175. doi: 10.1093/emboj/17.11.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelder C, Dunn MA, Choy B, Suh Y, Cassie C, Shim EY, Shin TH, Mello C, Seydoux G, Blackwell TK. Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1. Genes Dev. 1999;13:202–212. doi: 10.1101/gad.13.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackell TK, Walker A. Transcription mechanisms. In: WormBook, editor. The C elegans Research Community. Wormbook; 2006. [DOI] [Google Scholar]
- Bowerman B, Ingram MK, Hunter CP. The maternal par genes and the segregation of cell fate specification activities in early Caenorhabditis elegans embryos. Development. 1997;124:3815–3826. doi: 10.1242/dev.124.19.3815. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Colegrove-Otero LJ, Minshall N, Standart N. RNA-Binding Proteins in Early Development. Critical Reviews in Biochemistry and Molecular Biology. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- de Moor CH. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 1999;18:2294–2303. doi: 10.1093/emboj/18.8.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Dean KA, Aggarwal AK, Wharton RP. Translational repressors in Drosophila. Trends Genet. 2002;18:572–577. doi: 10.1016/s0168-9525(02)02792-0. [DOI] [PubMed] [Google Scholar]
- Detwiler MR, Reuben M, Li X, Rogers E, Lin R. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev Cell. 2001;1:187–199. doi: 10.1016/s1534-5807(01)00026-0. [DOI] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- Evans TC, Crittenden SL, Kodoyianni V, Kimble J. Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell. 1994;77:183–194. doi: 10.1016/0092-8674(94)90311-5. [DOI] [PubMed] [Google Scholar]
- Evans TC, Hunter CP. Translational control of maternal RNAs. In: WormBook, editor. The C elegans Research Community. Wormbook; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley BM, Pagano JM, Ryder SP. RNA target specificity of the embryonic cell fate determinant POS-1. RNA. 2008;14:2685–2697. doi: 10.1261/rna.1256708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley BM, Ryder SP. Regulation of maternal mRNAs in early development. Critical Reviews in Biochemistry and Molecular Biology. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]
- Farley BM, Ryder SP. POS-1 and GLD-1 repress glp-1 translation through a conserved binding-site cluster. Mol Biol Cell. 2012;23:4473–4483. doi: 10.1091/mbc.E12-03-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Maine E, Schedl T. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics. 1995;139:607–630. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Russel S, Ruvkun G. Functional Genomic Analysis of C. elegans Molting. PLoS Biol. 2005;3:1719–1733. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Ahkmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleland CL, Falls AT, Noraky J, Heiman MG, Yanik MF. Computer-Assisted Transgenesis of Caenorhabditis elegans for Deep Phenotyping. Genetics. 2015;201:39–46. doi: 10.1534/genetics.115.179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NN, Mootz DE, Walhout AJM, Vidal M, Hunter CP. MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development. 2002;129:747–759. doi: 10.1242/dev.129.3.747. [DOI] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annual Review of Biochemistry. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Kalchhauser I, Farley BM, Pauli S, Ryder SP, Ciosk R. FBF represses the Cip/Kip cell-cycle inhibitor CKI-2 to promote self-renewal of germline stem cells in C. elegans. EMBO J. 2011;30:3823–3829. doi: 10.1038/emboj.2011.263. http://www.nature.com/emboj/journal/v30/n18/full/emboj2011263a.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Karashima T, Sugimoto A, Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- Kaymak E, Ryder SP. RNA recognition by the Caenorhabditis elegans oocyte maturation determinant OMA-1. J Biol Chem. 2013;288:30463–30472. doi: 10.1074/jbc.M113.496547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ishidate T, Ghanta KS, Seth M, Conte D, Shirayama M, Mello CC. A Co-CRISPR Strategy for Efficient Genome Editing in Caenorhabditis elegans. Genetics. 2014;197:1069–1080. doi: 10.1534/genetics.114.166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M, Hartwieg E, Horvitz HR. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development. 1996;122:2579–2588. doi: 10.1242/dev.122.9.2579. [DOI] [PubMed] [Google Scholar]
- Lasko P. Cup-ling oskar RNA localization and translational control. J Cell Biol. 2003;163:1189–1191. doi: 10.1083/jcb.200311123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman JL, Jongens TA. Transcriptional silencing and translational control: key features of early germline development. Bioessays. 2003;25:326–335. doi: 10.1002/bies.10247. [DOI] [PubMed] [Google Scholar]
- Lin R. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev Biol. 2003;258:226–239. doi: 10.1016/s0012-1606(03)00119-2. [DOI] [PubMed] [Google Scholar]
- Lublin AL, Evans TC. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev Biol. 2007;303:635–649. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Marin VA, Evans TC. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development. 2003;130:2623–2632. doi: 10.1242/dev.00486. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Melton DA. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328:80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–3272. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Otori M, Karashima T, Yamamoto M. The Caenorhabditis elegans homologue of deleted in azoospermia is involved in the sperm/oocyte switch. Mol Biol Cell. 2006;17:3147–3155. doi: 10.1091/mbc.E05-11-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, Essien KI, Ryder SP. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc Natl Acad Sci. 2009;106:20252–20257. doi: 10.1073/pnas.0907916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, McCoig LM, Ryder SP. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem. 2007;282:8883–8894. doi: 10.1074/jbc.M700079200. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat Struct Mol Biol. 2004;11:20–28. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, Kruglyak L. A Novel Sperm-Delivered Toxin Causes Late-Stage Embryo Lethality and Transmission Ratio Distortion in C. elegans. Ed by. Laurence D Hurst. PLoS Biol. 2011;9:e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Jr, Mello CC. The C. elegans CSR-1 Argonaute Pathway Counteracts Epigenetic Silencing to Promote Germline Gene Expression. Dev Cell. 2013;27:656–663. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G. Mechanisms of translational control in early development. Curr Opin Genet Dev. 1996;6:555–561. doi: 10.1016/s0959-437x(96)80083-9. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Shimada M, Kawahara H, Doi H. Novel family of CCCH-type zinc-finger proteins, MOE-1, -2 and -3, participates in C. elegans oocyte maturation. Genes Cells. 2002;7:933–947. doi: 10.1046/j.1365-2443.2002.00570.x. [DOI] [PubMed] [Google Scholar]
- Simonet T, Dulermo R, Schott S, Palladino F. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev Biol. 2007;312:367–383. doi: 10.1016/j.ydbio.2007.09.035. [DOI] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annual Review of Biochemistry. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Spike CA, Coetzee D, Eichten C, Wang X, Hansen D, Greenstein D. The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of Caenorhabditis elegans oocytes. Genetics. 2014a;198:1535–1558. doi: 10.1534/genetics.114.168831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike CA, Coetzee D, Nishi Y, Guven-Ozkan T, Oldenbroek M, Yamamoto I, Lin R, Greenstein DI. Translational Control of the Oogenic Program by Components of OMA Ribonucleoprotein Particles in Caenorhabditis elegans. Genetics. 2014b;198:1513–1533. doi: 10.1534/genetics.114.168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS. “Masked” forms of mRNA. Curr Top Dev Biol. 1966;1:1–38. [PubMed] [Google Scholar]
- Stitzel ML, Cheng KCC, Seydoux G. Regulation of MBK-2/Dyrk kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr Biol. 2007;17:1545–1554. doi: 10.1016/j.cub.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316:407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Thompson B, Wickens M, Kimble J. 19 Translational Control in Development. Cold Spring Harbor Monograph Archive. 2007;48:507–544. [Google Scholar]
- Thyagarajan B, Blaszczak AG, Chandler KJ, Watts JL, Johnson WE, Graves BJ. ETS-4 is a transcriptional regulator of life span in Caenorhabditis elegans. PLoS Genet. 2010;6:e1001125. doi: 10.1371/journal.pgen.1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Ruggiu M, Cooke HJ. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res. 2001;29:2479–2483. doi: 10.1093/nar/29.12.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Boag PR, Blackwell TK. Transcription reactivation steps stimulated by oocyte maturation in C. elegans. Dev Biol. 2007;304:382–393. doi: 10.1016/j.ydbio.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein D, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends in Genetics. 2002:1–8. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wickens M, Goodwin EB, Kimble J. Translational control of developmental decisions. Cold Spring Harbor Laboratory Press; New York: 2000. pp. 295–370. [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 2010;30:533–545. doi: 10.1038/emboj.2010.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Li Y, Wang F, Noda NN, Zhang H. Differential function of the two Atg4 homologues in the aggrephagy pathway in Caenorhabditis elegans. J Biol Chem. 2012;287:29457–29467. doi: 10.1074/jbc.M112.365676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Bedet C, Robert VJP, Simonet T, Dunkelbarger S, Rakotomalala C, Soete G, Korswagen HC, Strome S, Palladino F. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc Natl Acad Sci. 2011;108:8305–8310. doi: 10.1073/pnas.1019290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Strome S. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics. 2001;159:1019–1029. doi: 10.1093/genetics/159.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]