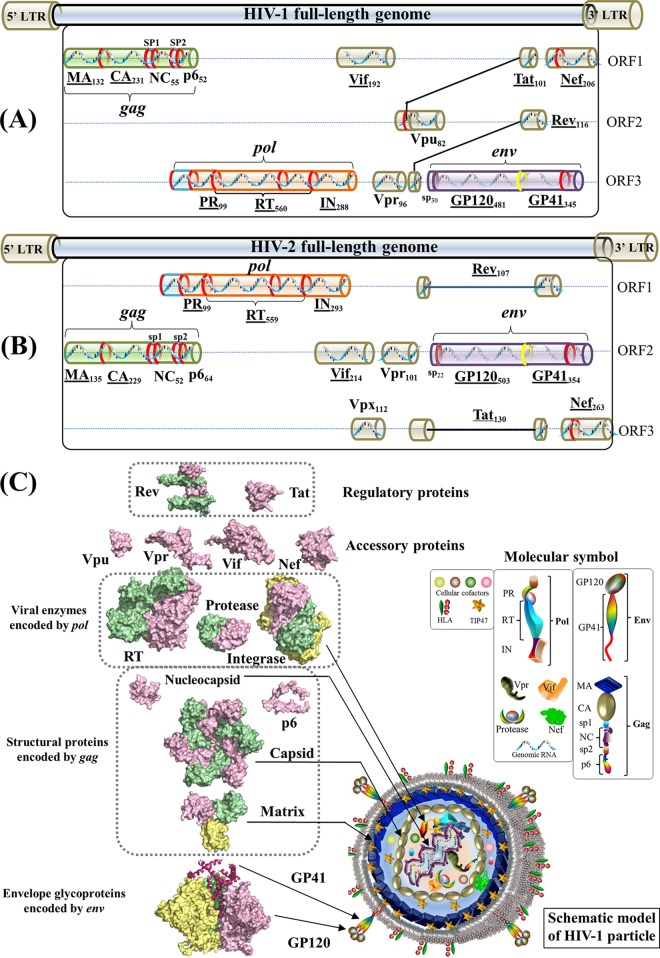
FIG 1.
Gene maps and protein structures of HIV-1 and HIV-2. (A) Schematic model of the HIV-1 full-length genome (reference strain HXB2). HIV protein names and amino acid lengths are shown beneath the colored protein regions in three open reading frames (ORFs). Eleven multimeric proteins have underlined names. In the gene map, red rings mark the locations where viral protease cleaves during viral maturation. In the env gene, a yellow ring shows the cleavage position of human proteases (e.g., furin) (594). The 5′ and 3′ long terminal regions (LTRs) are also indicated in the full-length genome. (Adapted from reference 44.) (B) Schematic model of the HIV-2 full-length genome (reference strain SIVmac239). (Adapted from reference 44.) (C) Surface representations of HIV-1 protein structures and schematic view of the HIV-1 particle. Surface representations of 15 HIV-1 protein structures are clustered according to their functional roles. HIV-1 monomeric proteins are shown in pink, and different subunits of multimeric proteins are distinguished with different colors (green, yellow, and red). HIV-1 protein structures are scaled precisely for a direct and intuitive comparison. At the bottom right, a schematic model of a mature viral particle is displayed, and the key shows protein annotations. Proteins in the schematic view are shown for illustration purposes; their structures and sizes here are not necessarily identical to the real protein structures and sizes. Additional information about HIV genomic reference sequences and natural polymorphisms is available online (see http://www.virusface.com/). (Adapted from reference 44.)