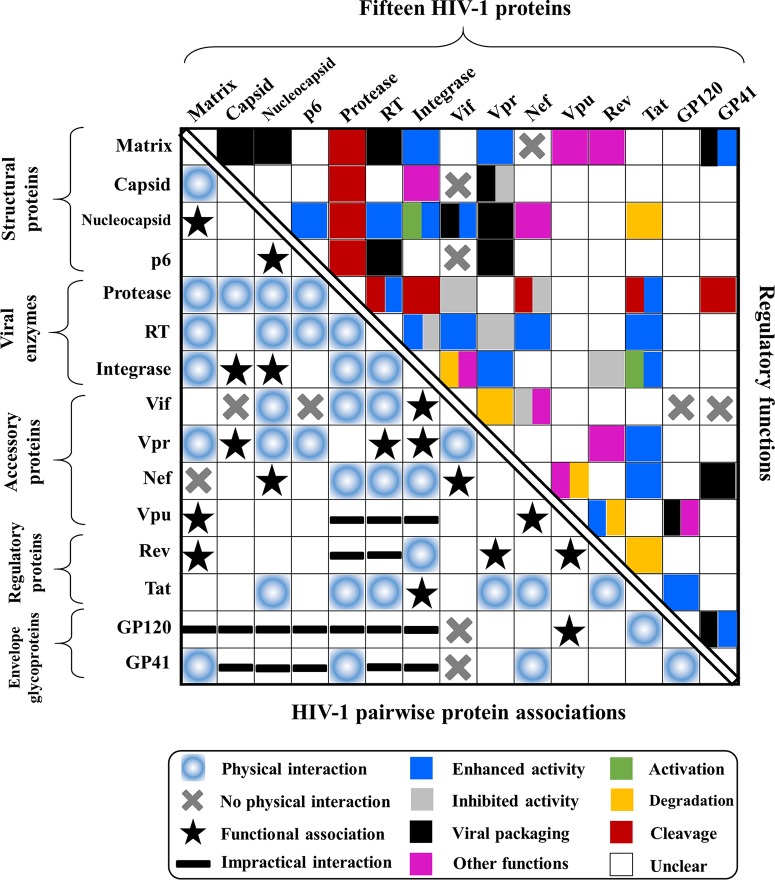
FIG 12.
Summary of possible HIV-1 pairwise protein associations. The names of 15 HIV-1 proteins are shown at the left and top. Five HIV-1 protein classes (envelope glycoproteins, regulatory proteins, accessory proteins, viral enzymes, and structural proteins) are shown on the left. The biological functions of HIV-1 pairwise protein associations are shown at the top right. Protein functions are mapped with different colors. If one protein association bears multiple functions, at most two major functions are annotated, and the mapped squares combine different colors inside. At the bottom is a summary of HIV-1 pairwise protein associations. Black stars indicate protein associations where two HIV-1 proteins are associated via a third molecule (Table 3), and black horizontal lines indicate impractical interactions between HIV-1 proteins, which are unlikely to take place during the HIV-1 life cycle (see the text for details). Empty squares without any mark inside imply that HIV protein associations or interactions remain unclear. Note that interactions involved with Gag and Env precursors are decomposed into individual protein interactions. For instance, the Gag-RT interaction (347) is decomposed into the matrix-RT and p6-RT interactions. The Gag-Vpr interaction leads to the NC-Vpr and p6-Vpr interactions. MatrixGag connecting with capsidGag in Gag results in the matrix-capsid interaction. Due to the limited number of reports on HIV-2, only information on HIV-1 protein associations is presented.