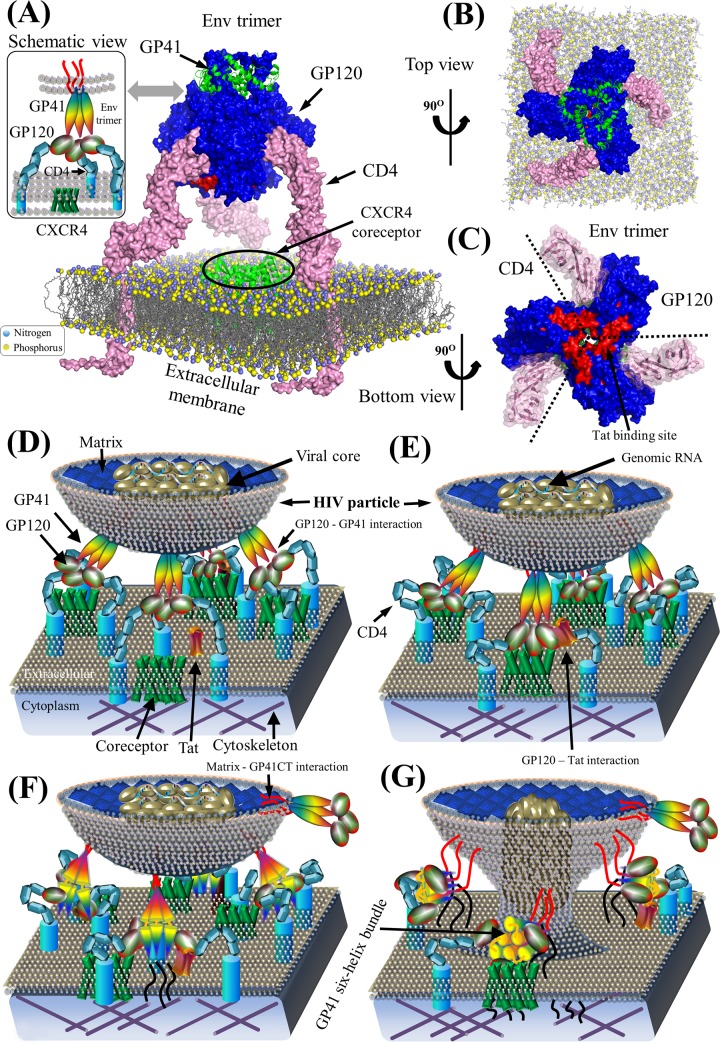
FIG 5.
Env structure complex and schematic model of HIV-1 pairwise protein interactions during viral entry. (A) Structural model of a prefusion HIV-1 Env spike associated with CD4 on the extracellular membrane. Surface representations of GP120, GP41, and CD4 proteins are shown in blue, green, and pink, respectively. Lipid bilayers of the extracellular membrane (606) are shown at the bottom, where nitrogen and phosphorus are indicated by blue and yellow spheres, respectively. The crystallized structure of the CXCR4 coreceptor in green is placed in the center across the extracellular membrane. Red areas on the GP120 surface illustrate the Tat-binding site (86, 89). Table S1 in the supplemental material provides a list of PDB accession numbers used for our structural visualization. PyMOL V1.7 visualization software was used (see http://www.pymol.org/). (B) Top view of a prefusion HIV-1 Env spike in complex with CD4 on the extracellular membrane. (C) Bottom view of the prefusion HIV-1 Env spike in complex with CD4. GP120 subunits within the trimeric Env spike bind to CD4. Red areas indicate Tat-binding sites. (E) Schematic view of the binding of Env to CD4 and coreceptors for viral attachment to the host membrane. GP120 on the mature virion surface interacts with CD4 to induce the aggregation of CD4 and chemokine coreceptors (e.g., CCR5 and CXCR4) (607, 608). Thereafter, GP120 binds to chemokine coreceptors on the host membrane. (F) Construction of GP41 six-helix bundles. Interactions between GP120 and chemokine coreceptors induce conformation rearrangements in Env spikes, which expose GP41 to construct the six-helix bundles (20). (G) Viral entry. GP41 six-helix bundles pull the extracellular membrane to create a fusion pore, which might consist of 1 to 7 Env spikes depending on divergent HIV strains (61, 62). The viral core in the HIV particle is then injected into the host cytoplasm by entering the newly created fusion pore. Note that protein shapes do not represent the exact protein structures, nor are the protein sizes to scale.