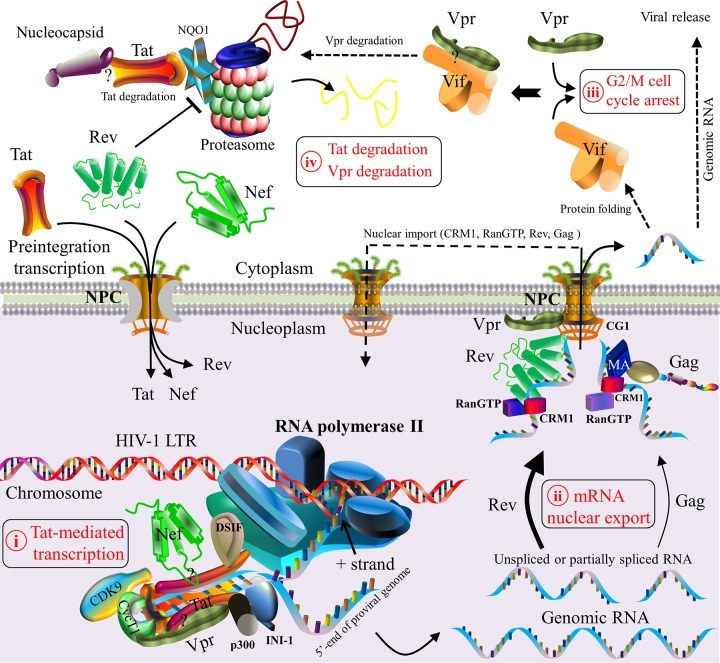
FIG 8.
Schematic model of HIV-1 protein interactions during viral transcription and translation. Five steps are described. (i) HIV-1 Tat initiates viral transcription by its interaction with the TAR of viral RNA, which is a regulatory element located downstream of the HIV-1 LTR (212). Tat subsequently recruits the subunits of the positive transcription elongation factor (e.g., cyclin T1 [CycT1] and cyclin-dependent kinase 9 [CDK9]) to construct a transcription complex (212). This complex activates the kinase CDK9 for the hyperphosphorylation of RNA polymerase II, which interacts with Tat and other host factors (e.g., DSIF [5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor] [614]) to produce viral genomic mRNAs (615, 616). Genomic mRNAs are spliced thereafter. Although many host proteins take part in viral transcription (23), only CDK9, cyclin T1, p300, DSIF, INI1, and RNA polymerase II are shown. (ii) Nuclear export of newly synthesized viral RNAs is accomplished by either Rev- or Gag-mediated pathways (239–242). In the former case, Rev recruits cellular factors (e.g., CRM1 and RanGTP) to export viral genomic RNAs as well as incompletely spliced and unspliced mRNAs from the nucleoplasm to the cytoplasm (30, 213). The binding of Vpr to the nuclear pore complex (NPC) is predominantly localized in the nuclear envelope of the nucleus (247, 248). In the latter case, RNA nuclear export is activated by the nuclear export signal (NES) of matrixGag, which interacts with CRM1 (214, 242). The nuclear export of viral RNA allows viral protein maturation in the cytoplasm and cellular compartments. CRM1, RanGTP, Rev, and Gag are then imported back to the nucleus (213, 214, 242). (iii) HIV-1 Vif and Vpr, both of which are expressed in the cytoplasm, can independently trigger G2/M cell cycle arrest (233). Vif may also interact with Vpr to mediate the degradation of Vpr and to reduce Vpr-induced cell cycle arrest (232). (iv) At the late stage of viral transcription, the proteasomal degradation of Tat is induced by viral nucleocapsid (228). Moreover, Rev induces Tat degradation by downregulating the expression level of the cellular protein NQO1 (216). Question marks indicate unclear interaction domains. Note that protein shapes do not represent the exact protein structures, nor are the protein sizes to scale.