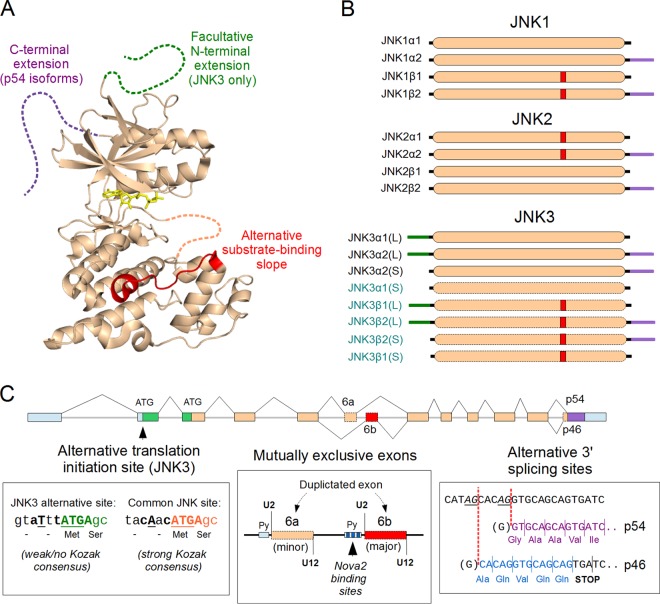
FIG 3.
Splice isoforms of the JNK1, JNK2, and JNK3 proteins. (A) The structure of JNKs. The generic structure of the JNK proteins is displayed in beige (represented by the crystal structure of JNK1β1; PDB ID 2XRW), and the variable regions (alternative splice isoforms) are highlighted in green, red, and magenta. The catalytic site of the kinase domain, ATP, is indicated in yellow. Regions that are unstructured or flexible are drawn with dotted lines. (B) All human JNK genes encode multiple splice isoforms. Apart from transcripts lacking a complete kinase domain (and therefore likely not yielding a functional protein), there are two variable regions for JNK1/2 and three for JNK3. All these alternative splicing products (as well as those resulting from alternative initiation with JNK3) combine freely and yield four isoforms for JNK1 and JNK2. For JNK3, there are 8 possible isoforms (including the longer [L] and shorter [S] N-terminal extensions), but only 3 isoforms have been characterized to date. However, mRNA sequences from databases (such as ENSEMBL) suggest that, like JNK1/2, JNK3 also contains the same alternative exons in its kinase domain. This hints at the existence of many more uncharacterized JNK3 isoforms (in blue). In the figure, the alternative segments structurally and evolutionarily corresponding to each other are labeled with the same colors: red, within the kinase domain; magenta, C-terminal flexible extension; green, N-terminal flexible extension. (C) Mechanisms of splice isoform generation in vertebrate JNK genes. The JNK3 gene has an upstream ATG codon, resulting in N-terminally extended proteins (green). However, this upstream initiation site has no Kozak consensus sequence, and so this is expected to result in “leaky scanning” by ribosomes, allowing the translational start to stochastically shift downstream to the site shared with all other JNK proteins. All vertebrate JNK genes have a duplicated exon (exon 6a [beige] and exon 6b [red]), where nonregulated splicing 6b is the preferred (major) exon. Their inclusion in the final transcript is mutually exclusive with each other because of the incompatibility of their U2- and U12-recognized splicing sites. Inclusion of the 6a exon depends on the suppression of exon 6b splicing, which can happen when the Nova2 protein binds to its polypyrimidine tract (Py) in JNK2. The ultimate splicing site is also variable, allowing for a 5-nucleotide shift. This results in a frameshift and an early stop codon in the short (p46) isoforms, while allowing the translation of the last exon in full in the case of the long (p54) isoforms. The sequences of the p46 (blue) and p54 (magenta) isoforms in the figure refer to JNK1. Note that the generic intron-exon pattern (colored to match the alternative protein sequences) shown at the top is not proportional to actual intron-exon sizes. The untranslated regions are displayed in light blue.