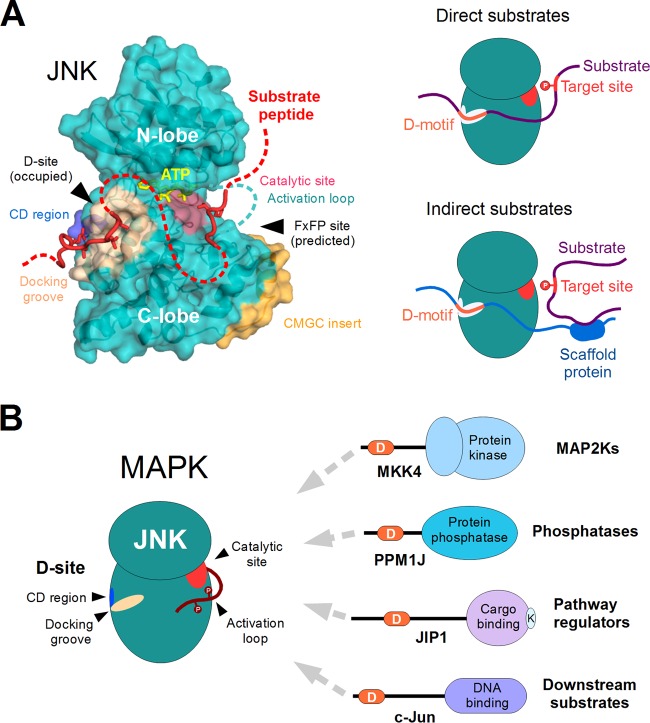
FIG 4.
Structural features and substrate recognition by JNKs. (A) JNK proteins are comprised of a single protein kinase domain (structure on the left). The docking site, consisting of the negatively charged CD region (blue) and the hydrophobic docking groove (beige), plays an important role in partner binding and recruitment of substrates (red). The phosphotransfer reaction from ATP (yellow) takes place at the opposite side of the kinase, where the catalytic residues (pink) are located. Apart from the docking site, the CMGC insert (orange) is also unique to the MAPKs and a few related protein kinases. This also harbors a docking surface called the FxFP site. Although known to be functional in other MAPKs, no FxFP site-dependent substrates have been identified for JNK. The figure is based on the complex of human JNK1 with a docking motif from NFAT4 (PDB ID 2XRW). The peptide chain modeled at the catalytic site is based on the DYRK1A-substrate complex (PDB ID 2WO6; DYRKs are closely related to MAPKs in structure as well as in substrate preference). The rest of the substrate, which is not associated with JNK, is indicated with a dotted red line. Together, the CD region and the docking groove form the major docking site (D-site) of JNK proteins and play a key role in substrate recruitment (shown on the right). The best-characterized substrate proteins either contain a linear motif capable of interacting with the D-site directly (direct substrates [top]) or interact with a third protein having such a motif through heterologous interactions (indirect substrates [bottom]). (B) JNKs bind most of their known partners by engaging a dedicated recruitment site (D-site) that is distinct from their catalytic site. The same docking site is used to interact with activator kinases (MAP2Ks) responsible for the phosphorylation of the JNK activation loop, with phosphatases that dephosphorylate the same residues, as well as with other proteins involved in the regulation of pathway through intracellular compartmentalization and multiprotein complex formation (i.e., scaffolds). Many substrates also utilize the same docking site to provide access to the kinase. Therefore, most partners of JNKs directly compete with each other for binding and access to the catalytic site. Abbreviations: D, docking motif; K, kinesin-binding motif of JIP1.