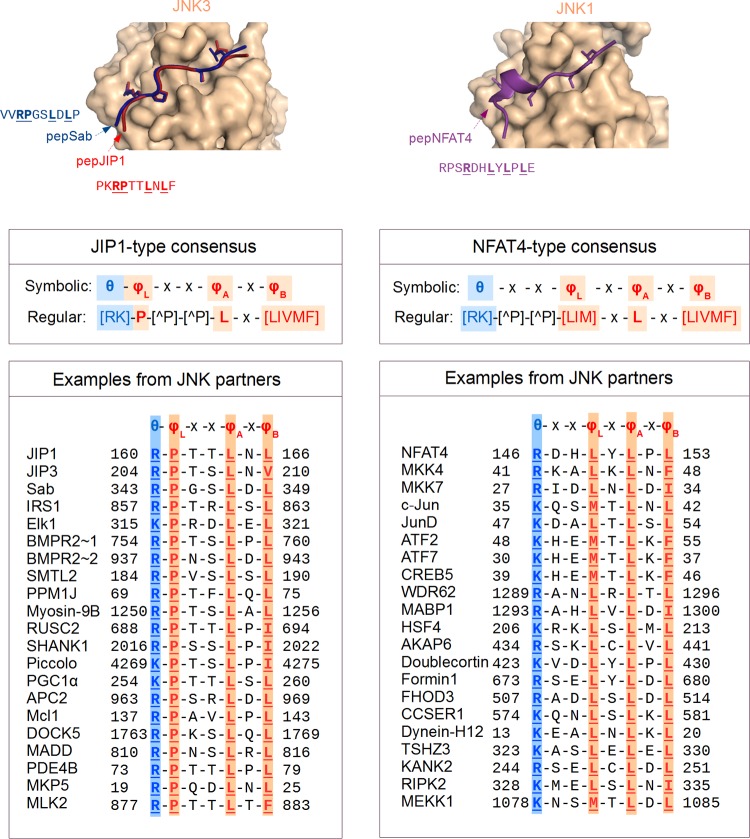
FIG 5.
The two main classes of D-motifs that interact with JNKs. Most of the known JNK-interacting D-motifs (located in diverse partners) belong to one of two distinct structural types, corresponding either to the JIP1 or to the NFAT4 consensus motifs (top). These two structural classes can be described with related, though different, consensus motifs (middle). Despite the differences, all these motifs bind to the same docking site. A large number of known JNK interactors, together with their evolutionarily closely related paralogs, harbor docking motifs showing sequence similarity to either the docking motif of JIP1 or to the docking motif of NFAT4 (bottom panels). Many of these docking motifs were characterized in in vitro experiments only, a few motifs do not satisfy the complete consensus, and some proteins (e.g., BMPR2, ATF2, ATF7, MKK7) contain more than one motif of the same or different type. (Structural panels were made by using crystal structures of JNK-peptide complexes: PDB IDs 4H39, 4H3B, and 2XRW for JNK3-pepJip1, JNK3-pepSab, and JNK1-pepNFAT4, respectively.)