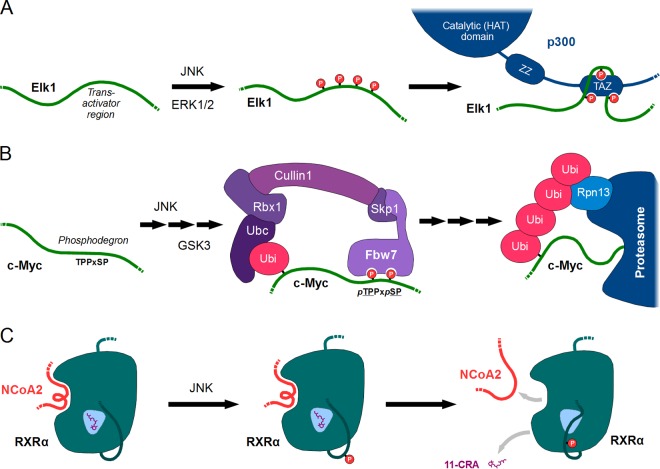
FIG 9.
Positive phospho-switches and allosteric switches controlled by JNK. (A) A simple positive switch acting on Elk1. Phosphorylation of the ETS transcription factor Elk1 by JNK1 (or ERK1/2) on multiple sites at its transactivation region creates a new linear motif. This protein-protein interaction motif binds to the histone acetyltransferase CBP/P300, likely through its second TAZ zinc finger domain. Thus, Elk1 can recruit chromatin-modifying enzymes that enhance transcription of its target genes. (B) A complex positive switch exemplified by phosphodegron systems. Several substrates of JNK, such as c-Myc, contain phosphodegron motifs. Multistep phosphorylation of such a motif (using GSK3) results in the recruitment of an E3 ubiquitin ligase complex containing the recognition subunit FBW7. Subsequent ubiquitinylation will then generate another new protein-protein interaction, this time with the lid of the proteasome. The result is usually the degradation of the ubiquitinylated protein, which in this sense is an inhibitory outcome (despite all protein-protein interactions being positive). (C) An allosteric switch on RXRα elicited by JNK-dependent phosphorylation of a regulatory loop. The retinoid X receptor α is an allosterically sensitive protein which only recruits the coactivator NCoA2 in its ligand (11-cis-retinoic acid [11-CRA])-bound state. The JNK-phosphorylated loop is directly adjacent to the ligand-binding pocket, and all available evidence suggests that it will bind back to the pocket when modified. The consequential distortion of the ligand-binding site would not only elicit release of 11-CRA, but also its coactivators, thus shutting down RXRα-dependent transcription.