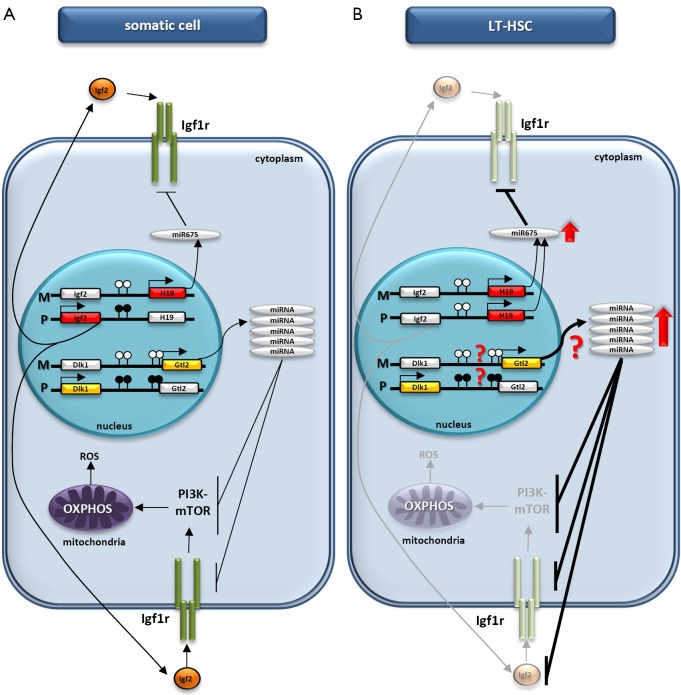
Figure 1.
Proposed regulation of HSC quiescence by paternally imprinted DMRs. (A) In somatic cells there is a balance between expression of the ncRNAs transcribed from the H19 and Gtl2 genes located on maternal (M) chromosomes and proteins encoded by the Igf2 and Dlk1 genes located on paternal (P) chromosomes. Methylated and unmethylated CpG sites in DMRs are shown in filled and open circles, respectively. (B) Due to changes in the methylation of DMRs in LT-HSCs, Igf2 expression is downregulated, whereas there is enrichment in the H19 ncRNA transcript, which leads to higher levels of miR-675 and a reduction in Igf1r expression. At the same time, enrichment in the Gtl2 ncRNA transcript with downstream miRNA cluster (for example, as a result of potential changes in methylation of the DMR within Dlk1–Gtl2) may result in increased levels of several miRNAs that target Igf1r, Igf2, and several components of the PI3K-mTOR pathway. The inhibition of Igf2/Igf1–Igf1r signaling as well as inhibition of the PI3K-mTOR pathway inhibits mitochondrial biogenesis and metabolic activity and increases the ROS level in LT-HSCs, leading to impaired hematopoiesis. The DMRs methylation state at the Dlk1–Gtl2 locus on the maternal and paternal chromosomes requires further study. HSC, hematopoietic stem cell; DMRs, differentially methylated regions; DMR, differentially methylated region; L-HSCs, long-term repopulating HSCs.