Summary
The Gram-positive pathogen Corynebacterium diphtheriae exports through the Sec apparatus many extracellular proteins that include the key virulence factors diphtheria toxin and the adhesive pili. How these proteins attain their native conformations after translocation as unfolded precursors remains elusive. The fact that the majority of these exported proteins contain multiple cysteine residues and that several membrane-bound oxidoreductases are encoded in the corynebacterial genome suggests the existence of an oxidative protein-folding pathway in this organism. Here we show that the shaft pilin SpaA harbors a disulfide bond in vivo and alanine substitution of these cysteines abrogates SpaA polymerization and leads to the secretion of degraded SpaA peptides. We then identified a thiol-disulfide oxidoreductase (MdbA), whose structure exhibits a conserved thioredoxin-like domain with a CPHC active site. Remarkably, deletion of mdbA results in a severe temperature-sensitive cell division phenotype. This mutant also fails to assemble pilus structures and is greatly defective in toxin production. Consistent with these defects, the ΔmdbA mutant is attenuated in a guinea pig model of diphtheritic toxemia. Given its diverse cellular functions in cell division, pilus assembly and toxin production, we propose that MdbA is a component of the general oxidative folding machine in C. diphtheriae.
Introduction
Corynebacterium diphtheriae is a Gram-positive actinobacterium that is infamous for its secretion of a highly potent toxin that causes diphtheria, a deadly disease for unvaccinated individuals (Rogers et al., 2011). Diphtheria toxin (DT) is a canonical A-B toxin comprised of a catalytic domain A, and transfer-mediating domain B. Following cleavage of the N-terminus by a host-derived endopeptididase, the domains remain linked by a single disulfide bond, which permits the release of domain A upon its entry into the reducing host cytoplasm (Collier, 2001). In addition, domain B contains another disulfide bond, which presumably aids in protein folding and stability (London, 1992). C. diphtheriae pathogenesis is also dependent on three types of adhesive pili that are assembled by a sortase-mediated mechanism (Ton-That and Schneewind, 2003; Mandlik et al., 2007). Similar to DT, all corynebacterial pilins possess a signal peptide sequence, apparently required for SecA-mediated translocation of unfolded protein precursors. A model of pilin export and assembly has been proposed for the archetype SpaA pili. According to this model (Mandlik et al., 2008; Reardon and Ton-That, 2013), pilin precursors are transported out of the cell by the Sec apparatus in an unfolded states, and then are retained within the extracytoplasmic face of the membrane by a C-terminal cell wall sorting signal (CWSS). This CWSS is required for sortase-catalyzed polymerization that joins individual subunits into a pilus polymer. The resulting polymers are ultimately linked to the bacterial cell wall via lipid II precursor. How pilins and DT are folded after translocation remains a fundamental question.
In Gram-negative bacteria, disulfide bond formation is critical for posttranslocational folding of proteins secreted into the oxidizing environment of the bacterial periplasm. In Escherichia coli, DsbA – the archetype of Gram-negative thiol-disulfide oxidoreductases (Bardwell et al., 1991) – oxidizes free thiol groups in nascent proteins emerging from the Sec translocon by donating a reactive disulfide bond within a catalytic CxxC motif (Kadokura and Beckwith, 2009). In turn, the DsbA active site is reduced and requires reoxidation by DsbB, a membrane-bound oxidoreductase (Bardwell et al., 1993; Missiakas et al., 1993). Recycling activities of the redox pair DsbA/DsbB is dependent on quinone, a component of the electron transport chain (Kobayashi and Ito, 1999). Importantly, because many Gram-negative virulence factors, such as flagella, adhesive pili and toxins, require disulfide bond formation, disulfide bond-forming machinery is key for virulence in a number of Gram-negative pathogens, including E. coli, Shigella flexneri, Pseudomonas aeruginosa, Bordetella pertussis, Neisseria meningitidis and Salmonella enterica (Heras et al., 2009).
By comparison, oxidative protein folding within Gram-positive bacteria has not been fully explored. The Gram-positive cell envelope is comprised of a single cytoplasmic membrane, hence called monoderm (Gupta, 1998), and so is considered devoid of a periplasmic space. Curiously, DsbA-like proteins have been identified in several Gram-positive organisms, but most are encoded in operons with specific substrates, unlike that in Gram-negative bacteria (Dorenbos et al., 2002; Meima et al., 2002; van der Kooi-Pol et al., 2012). In Bacillus subtilis, dsb-like genes are located within a competence gene cluster, and required for formation of the Com GC pilus (Meima et al., 2002). Only DsbA was found in Staphylococcus aureus (Dumoulin et al., 2005), and its function has also recently been implicated in stability of the ComGC pseudopilin (van der Kooi-Pol et al., 2012). More recently, a paralogous thiol-disulfide oxidoreductase in Streptococcus gordonii called SdbA has been linked to disulfide bond formation of autolysin AtlS (Davey et al., 2013). Unlike Firmicutes, Actinobacteria, including Corynebacterium, Streptomyces and Mycobacterium, secrete a large number of polypeptides with multiple Cys residues and encode thiol-oxidoreductase proteins (Daniels et al., 2010). Intriguingly, expression of a Mycobacterium tuberculosis oxidoreductase, annotated as Vitamin K epoxide reductase (VKOR), was able to rescue the defects of the E. coli dsbB mutant (Dutton et al., 2008). A DsbA-like protein has been reported in this organism (Chim et al., 2013; Premkumar et al., 2013). More recently, a disulfide bond forming machine has been identified in Actinomyces oris (Reardon-Robinson et al., 2015). These findings support the notion that Actinobacteria may utilize general oxidative protein folding in their exoplasm.
Here we describe a disulfide bond-forming machine for Sec-transported proteins in the actinobacterium C. diphtheriae. We demonstrate that the SpaA pilin of C. diphtheriae contains a disulfide bond that is essential for pilus assembly. Genetic and biochemical studies revealed that disulfide bond formation is catalyzed by a membrane-tethered thiol-disulfide oxidoreductase enzyme termed MdbA (mdb for monoderm disulfide bond-forming). Structure determination by X-ray crystallography showed the presence of a thioredoxin-like domain, typical of thiol-disulfide oxidoreductases. Importantly, we showed that the chaperone functions of MdbA are not limited to the pilins, as MdbA is essential for proper folding and stability of the diphtheria toxin. Furthermore, the mdbA mutant is defective in cell division at 37°C and also attenuated in virulence. Therefore, we propose that MdbA is a major disulfide bond-forming machine in C. diphtheriae that may serve as an important target for the development of antimicrobial therapies.
Results
Disulfide bond formation is required for pilus assembly in C. diphtheriae
Crystallization studies of the pilus shaft SpaA protein predicted that the C-terminal IgG-like domain contains a disulfide bond formed between cysteine (Cys) residues C383 and C443 (Kang et al., 2009) (see Fig. 1A). To probe the presence of this bond in vivo, we used methoxypolyethylene glycol-maleimide (Mal-PEG), a 2-KDa alkylating agent that forms stable thioether bonds with free sulfhydryl groups (Makmura et al., 2001), which can be detected by a change of protein mobility using SDS-PAGE. SpaA monomers were collected from the culture medium of a C. diphtheriae mutant (HT3) that secretes SpaA monomers (Swaminathan et al., 2007), trichloroacetic acid (TCA) precipitated and then acetone washed. The resulting samples were resuspended in buffer with or without DTT, and subsequently incubated with Mal-PEG. After removal of Mal-PEG by TCA precipitation, SpaA was detected by Western blotting using SpaA specific antibodies (α-SpaA). In the absence of DTT, Mal-PEG treatment did not alter migration of SpaA monomers (Fig. 1B; lanes 1 and 3). In contrast, reduction of SpaA with DTT prior to Mal-PEG treatment resulted in an upshift of the SpaA band (Fig. 1B; last lane), indicating that the pilin contains a disulfide bond in vivo.
Fig. 1.
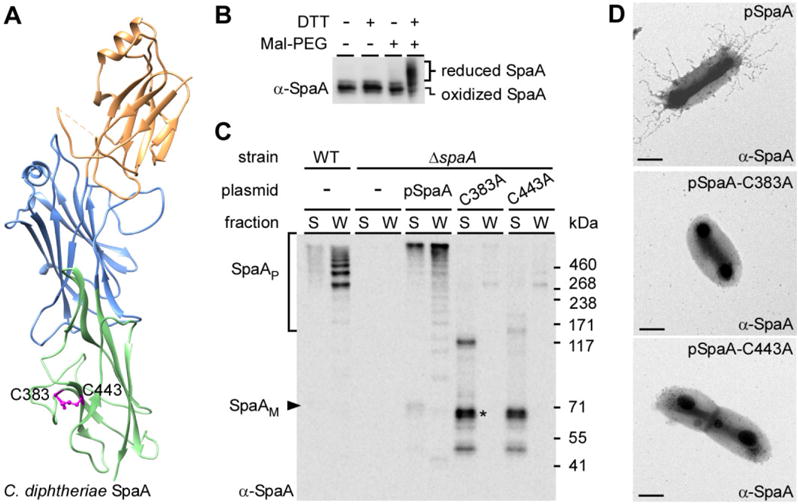
Requirement of disulfide bond formation in pilus assembly.
A. Cysteine residues C383 and C443 are predicted to form the C-terminal disulfide bond of the SpaA crystal structure (PDB:3HR6) [25].
B. SpaA monomers were isolated and treated or mock-treated with DTT, followed by Mal-PEG alkylation. The protein samples were immunoblotted with antibodies against SpaA (α-SpaA). Reduced and oxidized forms of SpaA are indicated.
C. Culture medium (S) and cell wall (W) fractions were collected from corynebacterial cultures. Equivalent protein samples obtained by TCA precipitation were immunoblotted with α-SpaA. Monomeric and polymeric forms of SpaA, as well as molecular mass markers (kDa) are indicated. SpaA degraded products are denoted by an asterisk.
D. Cells of corynebacterial strains overexpressing wild-type SpaA (pSpaA) or its isogenic mutants (C383A or C443A) were immobilized on formvar carbon-coated nickel grids, and stained with α-SpaA, followed by anti-rabbit IgG conjugated to gold particles. The samples were then stained with 1% uranyl acetate and viewed by a transmission electron microscope. Scale bars indicate 0.2 μm.
We next examined whether the formation of this C-terminal disulfide bond is required for pilus assembly. A plasmid expressing SpaA harboring alanine-substitutions of C383 or C443 was introduced into a spaA deletion mutant (ΔspaA). Corynebacterial cells were subject to cell fractionation and protein samples were analyzed by Western blotting with (α-SpaA) (see Methods). Compared with cells expressing wild-type SpaA (Fig. 1C; lanes WT and pSpaA), no pilus polymers (marked P) were detected in the cell wall (W) fractions of the Cys-to-Ala mutants; instead, low-molecular-weight species of SpaA were detected in the culture medium (S) fractions (Fig. 1C; last four lanes). Cell surface assembly of pili was also examined by immunogold electron microscopy (IEM). Corynebacteria were immobilized on carbon-coated nickel grids, blotted with α-SpaA, followed by IgG antibodies conjugated to gold particles. Consistently, no pili were observed on the surface of C383A or C443A mutants (Fig. 1D). Together, these results demonstrate that the SpaA C383-C443 linkage is essential for pilus assembly.
Identification of a thiol-disulfide oxidoreductase in C. diphtheriae by X-ray crystallization
The C. diphtheriae genome encodes several putative membrane-bound oxidoreductases, including DIP0397, DIP1880 and DIP0411, that may participate in oxidative protein folding. DIP0411 is a proposed DsbF homolog and its crystal structure reveals a thioredoxin-like domain with a conserved catalytic CxxC motif (Um et al., 2014). However, its localization with genes encoding c-type cytochrome synthesis components suggests it may be involved with energy production rather than protein folding (Kanehisa and Goto, 2000). DIP0397 is predicted to encode a 32-kDa membrane-bound protein with a CPFC motif. A deletion of the gene encoding DIP0397 produced no detectable phenotypes, so it was not studied further (data not shown). Finally, DIP1880, which we renamed MdbA (mdb for monoderm disulfide bond-forming), is predicted to encode a 27-kDa transmembrane protein that harbors a CPHC motif. Cell fractionation and immunoblotting with antibodies against MdbA (α-MdbA) revealed that the protein is membrane localized (see Fig. 3A). We have recently shown that DIP1880 has thiol-oxidoreductase activities in vitro (Reardon-Robinson et al., 2015).
Fig. 3.
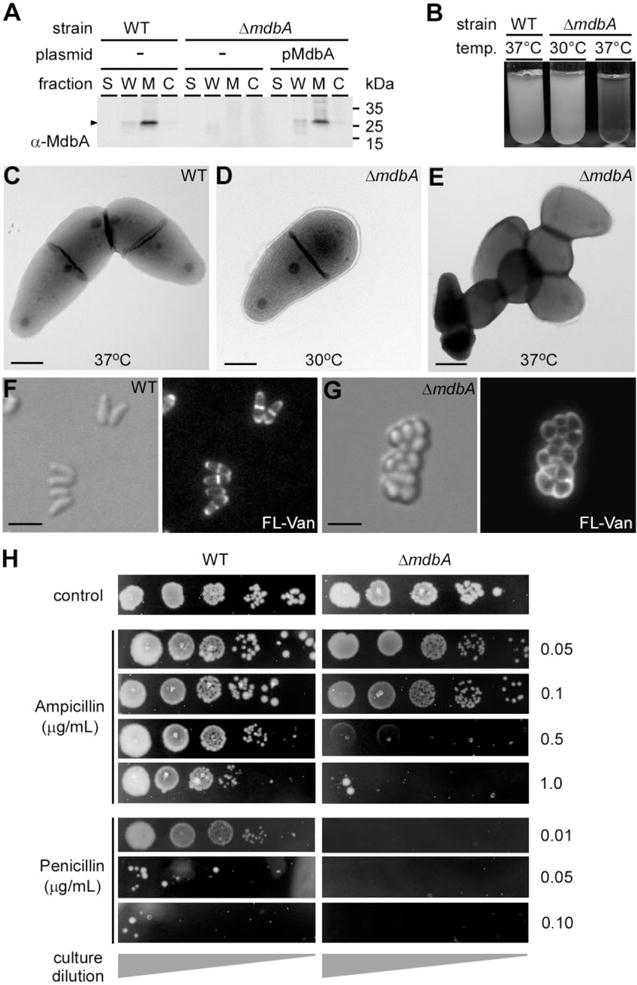
The C. diphtheriae ΔmdbA mutant is defective in cell growth and division.
A. Culture medium (S), cell wall (W), membrane (M) and cytoplasmic (C) fractions were collected from corynebacterial cultures. Protein samples were immunoblotted with antibodies against MdbA (α-MdbA).
B. Single colonies of the C. diphtheriae wild-type and mdbA mutant strains were inoculated in broth cultures overnight at 37°C or 30°C.
C–E. Stationary cultures of C. diphtheriae grown at 30°C were diluted in fresh media, and then incubated at 30°C or 37°C for 5 h. Harvested cells were immobilized on nickel grids, stained with 1% uranyl acetate and viewed by a transmission electron microscope. Scale bars indicate 0.5 μm.
F–G. Overnight cultures of C. diphtheriae grown at 30°C were diluted into fresh media, and then incubated at 37°C until wild-type cells reached log phase. A 1:1 mixture of vancomycin and fluorescent vancomycin (Van-FL) was added to cultures, and incubated for 10 min. The cells were placed directly on agar pads, and viewed by a fluorescence microscope. Scale bars indicate 2.5 μm.
H. Overnight cultures of C. diphtheriae grown at 30°C were diluted (10−3–10−7) and spotted on HIA plates containing various concentrations of ampicillin or penicillin, and incubated at 30°C for 48 h.
To reveal the structural identity of MdbA, we purified the recombinant protein from E. coli for X-ray crystallization using seleno-methionine single-wavelength anomalous diffraction (SAD). The reduced form of MdbAwas refined to 1.77-Å resolution with R-work and R-free factors equal to 16.9% and 21.0% respectively (Tables 1 and 2). The structure harbors a thioredoxin-like domain and an extended α-helical domain (Fig. 2A), typical of the DsbA protein family (Martin et al., 1993; Shouldice et al., 2011). The thioredoxin-like domain is composed of a 6-strand β-sheet in an order of β0↓- β1↑- β3↓- β2↓- β4↑- β5↓ and 2 flanking α helices, H1 and H7, based on a previous secondary structure numbering system (Martin et al., 1993). The β-sheet also includes an additional β0 strand at the N-terminus, presumably as a result of different positioning and/or higher completeness of the MdbA N-terminus, as compared with other Gram-positive DsbA-like structures (Fig. S1). The α-helical domain consists of 6 α-helices (H2–H6 and H3*) and two 310 helices (Fig. 2A and S1). Unlike other DsbA-like proteins, the C. diphtheriae MdbA structure contains a unique secondary structure element, which is the short α-helix H3* (residues 163–166) right after helix H3. The CPHC active-site motif (residues 91–94) resides on the N-terminal end of helix H1 (Fig. 2B). Significantly, the MdbA structure also harbors a conserved cis-Pro loop (residues S221 and P222) characteristically found in the TRX-related proteins (Ren et al., 2009). Situated on the loop between H6 α-helix and β5 strand, the cis-Pro element interacts with the CPHC motif via a hydrogen bond formed between S221 and C91 (3.07 Å distance, Fig. 2B).
Table 1.
Crystal data collection statistics.
| X-ray wavelength (Å) | 0.9792 | |
| Space group | P 21 | |
| Unit cell dimensions | a = 61.5 Å, | |
| b = 76.6 Å, | ||
| c = 72.6 Å, | ||
| α = γ = 90°, | ||
| β = 90.2° | ||
| Resolutiona (Å) | 33.9 – 1.77 | (1.80 – 1.77) |
| No. of unique reflections | 65737 | (3243) |
| Completeness (%) | 99.9 | (99.4) |
| R-merge | 0.078 | (0.62) |
| CC1/2 (Å2) | – | (0.75) |
| I/σ | 9.1 | (2.02) |
| Redundancy | 4.8 | (4.0) |
| Molecules per asymmetric unit | 4 | |
| No. of protein residues | 832 |
Numbers in parenthesis are shown for the highest resolution shell.
Table 2.
Structure refinement statistics.
| Resolution range (Å) | 34.0 – 1.77 | (1.82 – 1.77) |
| Reflections | 65 716 | (4 775) |
| σ cutoff | None | |
| R-value (all) (%) | 16.42 | |
| R-value (R-work) (%) | 16.19 | (26.9) |
| Free R-value (%) | 20.98 | (28.1) |
| Rms deviations from ideal geometry | ||
| Bond length (Å) | 0.013 | |
| Angle (degrees) | 1.50 | |
| Chiral (Å) | 0.093 | |
| No. of atoms | ||
| Protein | 6 357 | |
| Water | 399 | |
| Mean B-factor (Å2) | ||
| All atoms | 33.6 | |
| Protein atoms | 33.5 | |
| Protein main chain | 31.7 | |
| Protein side chain | 35.2 | |
| Water | 33.8 | |
| Molprobity Ramachandran plot statistics | ||
| Residues in favored regions (%) | 97.6 | |
| Residues in allowed regions (%) | 99.8 | |
| Residues in disallowed region (%) | 0.2 | |
Fig. 2.
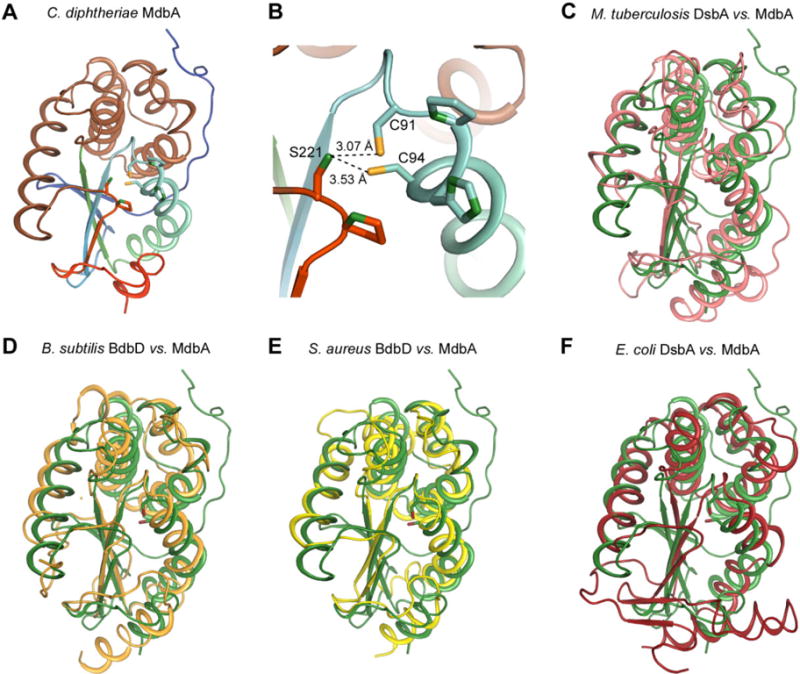
Structural analysis of the C. diphtheriae disulfide bond-forming protein MdbA.
A. The C. diphtheriae MdbA crystal structure (residues 43–244), solved to a 1.77-Å resolution, possesses a thioredoxin-like domain (rainbow colors) and an alpha-helical domain (light brown).
B. The MdbA active site is comprised of C91, P92, H93 and C94. The Sγ group in the two cysteine residues forms a hydrogen bond with S221 of the Cis-Pro element.
C–F. The MdbA structure (green) was aligned with M. tuberculosis DsbA (C; PDB: 4K6X), B. subtilis BdbB (D; PDB:3EU3), S. aureus DsbA (E; PDB:3BCI) and E. coli DsbA (F; PDB:1A2L).
Based on DALI server analysis (Holm and Rosenstrom, 2010), the MdbA structure is closely related to the crystal structure of Rv2969c, a DsbA-like protein from M. tuberculosis (PDB:4K6X) (Premkumar et al., 2013) (Fig. 2C), with Z-score and RMSD of 19.2 and 2.3, respectively, for 199 residues. Other similar structures include from B. subtilis BdbD (PDB: 3EU3) (Crow et al., 2009) and S. aureus DsbA (PDB: 3BCI) (Heras et al., 2008) (Fig. 2D and E). On the other hand, the E. coli DsbA structure (PDB:1A2L) (Martin et al., 1993) (Fig. 2F) is farther down on the DALI homolog list with Z-score and RMSD values equal to 11.4 and 3.7 respectively. Notably, the β-sheet topology of the C. diphtheriae MdbA structure is different from the E. coli DsbA and most of DsbA-family proteins of Gram-negative bacteria – the β-strand order of 0-1-3-2-4-5 compared with 3-2-4-5-1 in E. coli. Altogether, the results support that the C. diphtheriae MdbA is a thiol-disulfide oxidoreductase.
Essential roles of C. diphtheriae MdbA in cell division, pilus assembly, toxin production and disulfide bond formation
To examine the role of C. diphtheriae MdbA in vivo, we generated a nonpolar deletion mutant of mdbA. Strikingly, excision of this gene resulted in a severe growth defect at 37°C, while growth at 30°C was similar to that of the wild-type strain (Fig. 3B). When examined by electron microscopy, the ΔmdbA mutant was indistinguishable from wild-type at 30°C. However, when shifted to 37°C, ΔmdbA became chained, clumped and coccoid in appearance (Fig. 3C–E). To examine cell growth, we labeled corynebacteria with fluorescent vancomycin to visualize nascent peptidoglycan insertion by fluorescence microscopy. Fluorescence signal was detected at the septum and cell poles of the wild-type strain, consistent with apical growth of C. diphtheriae (Umeda and Amako, 1983); however, fluorescence signal was defused throughout the cell envelope of the ΔmdbA mutant (Fig. 3F and G). To probe a potential defect in cell wall synthesis, we tested C. diphtheriae susceptibility to β-lactams ampicillin and penicillin. Remarkably, ΔmdbA growth was inhibited at lower concentrations of antibiotics than WT (Fig. 3H). Altogether, the data suggest that mdbA is required for proper growth and division.
As pilus assembly is coupled with cell growth, pilus polymerization within the ΔmdbA mutant was examined by Western blotting using culture medium and cell wall fractions of bacteria grown at 30°C. Compared with wild-type corynebacteria, the ΔmdbA mutant did not produce any SpaA polymers, but complementation of this mutant with plasmid-borne copies of mdbA rescued this defect (Fig. 4A). Significantly, complementation was dependent on an intact CxxC active as alanine-substitution of MdbA C94 abrogated pilus assembly (see Fig. 5B).
Fig. 4.
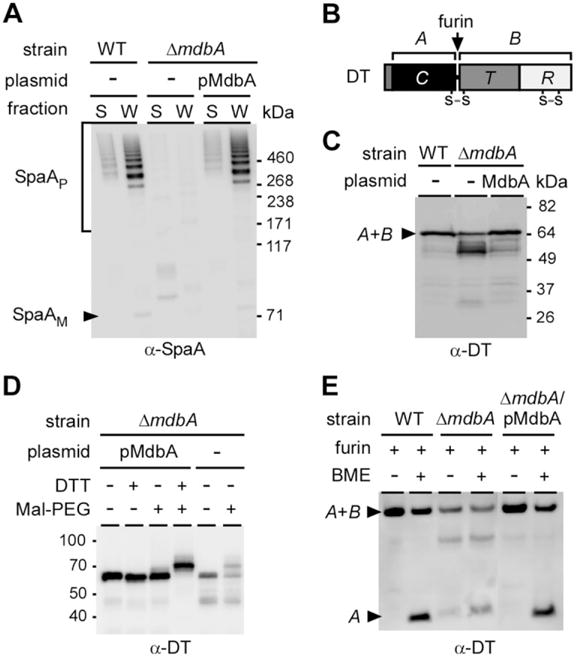
Requirement of the disulfide bond-forming machine MdbA for pilus assembly and diphtheria toxin (DT) production in C. diphtheriae.
A. Protein samples obtained from C. diphtheriae S and W fractions were separated on a 3–12% gradient gel and immunoblotted with α-SpaA.
B. A graphic presentation of DT highlights its two disulfide bonds, with the first one encircling domains A and B. A second disulfide bond is located at the B domain.
C. DT production was induced by addition of the iron chelator EDDA to culture media. Secreted DT was separated from cells by centrifugation and the protein samples were analyzed by Western blotting with monoclonal antibodies against domain A (α-DT).
D. DT collected from the culture medium of C. diphtheriae strains expressing or lacking MdbA was treated or mock-treated with DTT, followed by Mal-PEG alkylation. The protein samples were analyzed by immunoblotting with α-DT.
E. DT was collected from culture media, and concentrated by filter centrifugation. The protein was then treated with furin for 2 h at 37°C, boiled in SDS with or without β-mercaptoethanol, separated with a 12% Tris-Glycine gel and immunoblotted with α-DT. DT A and B fragments are indicated. Molecular mass markers are shown.
Fig. 5.
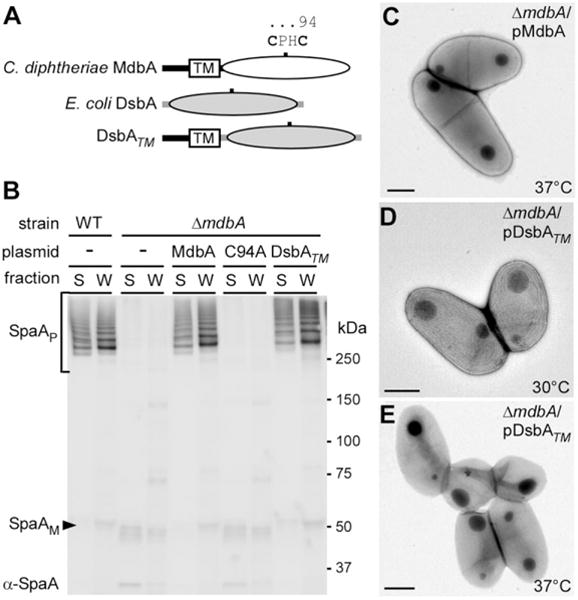
Conservation of disulfide bond-forming machinery.
A. A graphical representation of the primary sequences of C. diphtheriae MdbA and E. coli DsbA. The catalytic CPHC of MdbA is highlighted. To express E. coli DsbA in C. diphtheriae, the N-terminal signal peptide was swapped with the signal peptide and transmembrane anchor of MdbA.
B. Protein samples obtained from C. diphtheriae S and W fractions were separated on a 3–12% gradient gel and immunoblotted with α-SpaA.
C–E. Stationary cultures of C. diphtheriae grown at 30°C were diluted in fresh media, and then incubated at 30°C or 37°C for 5 h. Harvested cells were immobilized on nickel grids, stained with 1% uranyl acetate and viewed by a transmission electron microscope. Scale bars indicate 0.5 μm.
By bioinformatics analysis (Kanehisa and Goto, 2000), we found that more than 60% of proteins secreted by C. diphtheriae contain two or more cysteine residues. Therefore, we hypothesized that MdbA targets an array of substrates. To investigate this, we chose diphtheria toxin (DT) as a nonpilus protein model. DT is a protein synthesis inhibitor that belongs to the A-B family of toxins. Secreted by C. diphtheriae when bacteria encounter low iron, DT contains two disulfide bonds; the first disulfide bond loops the active domain A and the binding domain B, and is essential for toxin function (Boquet and Pappenheimer, 1976) (Fig. 4B). DT production was induced by adding a metal chelator to C. diphtheriae cultures, isolated by centrifugation, and then TCA precipitated. DT was then detected by Western blotting using monoclonal antibodies against the A fragment of the toxin (α-DT). DT from the wild-type strain was detected as a single band running at approximately 60 kDa as predicted (Fig. 4C). Deletion of mdbA, however, resulted in the production of multiple bands; the intensity of the band seen in wild-type cells is reduced with the concomitant accumulation of a low-molecular weight product(s) (Fig. 4C). In the absence of mdbA, this result suggested that DT is prone to degradation. Complementation of this mutant by wild-type mdbA rescued normal production of this toxin (Fig. 4C).
To test if the observed phenotype was caused by the failure to form disulfide bonds, we again turned to alkylation. Harvested DT was suspended in buffer with or without DTT, treated with Mal-PEG, and detected by Western blotting. DT obtained from wild-type cells showed no change in protein migration unless the sample was first treated with reducing DTT (Fig. 4D; first four lanes), indicating that secreted DT is oxidized. However, DT released by the ΔmdbA mutant was readily modified by Mal-PEG, demonstrating that it was released with reduced Cys residues (Fig. 4D, last two lanes). Note that the level of secreted DT in the ΔmdbA mutant was significantly reduced, consistent with the observation in Fig. 4C that DT is more susceptible to degradation when mdbA is absent.
The failure of the ΔmdbA mutant to form disulfide bonds in DT was also shown by examining the N-terminal disulfide bond that connects theAand B domains (Fig. 4E). Harvested DT was treated with furin, a protein that cleaves an Arg-XX-Arg motif between the A and B domains. Upon cleavage, the A and B domains are linked by a single disulfide linkage in the N-terminus. Following furin treatment, the samples were incubated with the reducing agent β-mercaptoethanol (BME) to separate the domains. In the sample obtained from wild-type cells, release of the A domain was detected only after BME treatment with BME. However, the release of the A domain in the sample obtained from ΔmdbA was detected immediately after furin cleavage, indicating that the disulfide bond between the domains was not formed. Expression of MdbA in the ΔmdbA mutant restored the wild-type pattern (Fig. 4E). Collectively, these results support the view that MdbA is required for disulfide bond formation within pilus and nonpilus virulence factors in C. diphtheriae.
Conservation of oxidative protein folding machinery
As MdbA is more closely related to M. tuberculosis (Mtb) DsbA, it is logical to determine if expression of Mtb DsbA would rescue the defects of the C. diphtheriae ΔmdbA mutant. However, the effort failed after several attempts to clone the Mtb dsbA gene in C. diphtheriae, possibly due to codon specificity. We then cloned the E. coli DsbA containing the transmembrane domain of MdbA into the C. diphtheriae ΔmdbA mutant (Fig. 5A; DsbATM). Pilus assembly was assessed by Western blotting as described in Fig. 4A with protein samples collected from the recombinant DsbA strain grown at 30°C. Remarkably, the ΔmdbA mutant expressing the membrane-tethered E. coli DsbA produced SpaA pilus polymers at the levels similar to that of the wild-type and MdbA complementing strains (Fig. 5B). However, this strain still exhibited slight cell morphology defects at 37°C (Fig. 5C–E), suggesting that the E. coli DsbA enzyme might not be fully functional.
Requirement of the disulfide bond-forming machine MdbA for C. diphtheriae virulence
As C. diphtheriae MdbA is required for the assembly of SpaA-type pili, as well as the production DT, we hypothesized that MdbA is an important virulence factor. To test this, we employed a guinea pig model of diphtheritic toxemia (Young and Mood, 1945; Pappenheimer et al., 1957). In this experiment, groups of six animals were injected via the intraperitoneal route (IP) with 2.5 × 107 CFU of the wild-type, Δtox, ΔspaA-I or ΔmdbA strain, and animal survival was monitored over 7 days (Fig. 6). Within 2 days, 80% of guinea pigs inoculated with wild-type corynebacteria succumbed to infection. The Δtox mutant, which does not produce diphtheria toxin, did not cause a lethal infection as expected. Remarkably, strains devoid of MdbA or all pili were significantly attenuated as well. There was little or no significant difference between the Δtox mutant strain and the ΔmdbA mutant or the ΔspaA-I mutant. These data demonstrate that MdbA is an important virulence factor for C. diphtheriae.
Fig. 6.
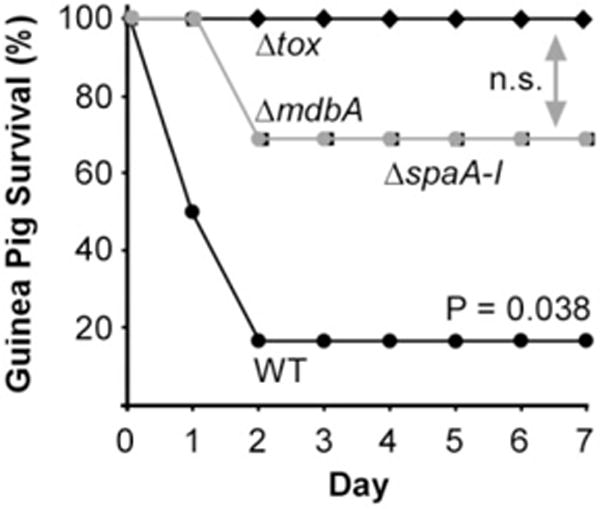
Requirement of MdbA for bacterial virulence. Six 4-week-old guinea pigs were infected via IP with ~ 2.5 × 107 corynebacteria of the wild-type (filled circles), pilus-less mutant (filled squares), toxin-less mutant (filled diamonds) or ΔmdbA mutant (gray circles) strain. Animal survival was monitored for 7 days. Statistical significance was analyzed by a paired t-test using GraphPad; n.s. denotes not significant.
Discussion
It is well known that oxidative protein folding in Gram-negative bacteria occurs in the oxidizing environment of their periplasmic space. Due to the lack of an outer membrane, however, the Gram-positive bacteria are generally not considered to possess an oxidative periplasm. Therefore, whether an oxidative protein-folding pathway exists in monoderm bacteria is an outstanding question. We report here our systematic investigation of a disulfide bond-forming enzyme MdbA that may serve as a general oxidative folding machine to fold exported proteins in C. diphtheriae.
To search for an oxidative folding machine in this organism, we employed two model exported proteins – SpaA and the diphtheria toxin, which are respectively predicted to and known to contain disulfide bonds. SpaA, the major subunit of the SpaA-type pili, is predicted to harbor a single disulfide bond between the residues C383 and C443 in the C-terminus. We confirmed the formation of this linkage in vivo using an alkylation assay (Fig. 1). Importantly, Cys-to-Ala mutations abolished pilus assembly, indicating that this linkage is essential for SpaA structure-function. We subsequently went on to identify the C. diphtheriae MdbA as a potential disulfide bond-forming enzyme through bioinformatics analysis. To determine if MdbA is indeed a thiol-oxidoreductase, we solved the 3-D structure of MdbA by X-ray crystallography. Like numerous thiol-oxidoreductases, C. diphtheriae MdbA has a signature thioredoxin-like domain and an extended α-helix (Fig. 2). Importantly, MdbA Cys residues are found within the CXXC catalytic site that interacts with a conserved cis-Pro loop. Comparisons of C. diphtheriae MdbA structure with other thiol-oxidoreductases revealed that it is more related to the mycobacterial DsbA than a structure solved for the well-known disulfide bond forming protein in E. coli.
Importantly, unlike Dsb proteins of E. coli, C. diphtheriae MdbA is a membrane-tethered enzyme (Fig. 3). We suspect that this may be an adaptation to the absence of an enclosed periplasmic compartment as E. coli dsbA rescued ΔmdbA phenotypes only if it was fused to a membrane anchor (Fig. 5B; data not shown). Remarkably, although the C. diphtheriae mdbA deletion mutant divided normally at 30°C, the cells exhibited a severe growth defect at 37°C. Electron microscopy revealed that the normally rod-shaped bacteria displayed chained morphology, and a coccoid-like appearance when shifted to the nonpermissive temperature. It is noteworthy that a previous study found that the penicillin-binding proteins (PBPs) expressed by Corynebacterium glutamicum were important for proper cell growth and division (Valbuena et al., 2007). Deletion of genes encoding these proteins caused similar morphological phenotypes observed with ΔmdbA. Not surprisingly, surveys of the C. diphtheriae genome revealed multiple PBPs with multiple Cys residues including PBP1a, PBP1b, PBP2a and PBP2b. PBP1b and PBP2b, which are synthetically lethal in C. glutamicum (Valbuena et al., 2007), possess 9 and 4 Cys residues respectively. We suspect that, similar to SpaA and DT, PBPs require oxidative folding. These proteins likely fail to fold properly in the absence of mdbA, thereby becoming nonfunctional and/or susceptible to proteolysis. In support of this, C. diphtheriae ΔmdbA was much more susceptible to antibiotics that target PBP function (Fig. 3). We speculate that at lower temperatures, exogenous sources of oxidation may fold enough PBPs to maintain proper cell growth. However, at 37°C, residual PBPs may not be sufficient to meet the demands of rapid cellular replication, and as a consequence, these bacteria eventually stop dividing. The role of MdbA in bacterial division will be the focus of future studies.
Importantly, we were able to establish the essential function of oxidative protein folding using two different model substrates. Deletion of mdbA abrogates pilus assembly (Fig. 4), and this is consistent with a defect in pilus polymerization when the C-terminal SpaA disulfide bond is disrupted (Fig. 1). These data demonstrate that the thiol-oxidoreductase MdbA machine is a required component of the pilus assembly pathway. Interestingly, although the C. diphtheriae MdbA structure is less similar than the E. coli DsbA, ectopic expression of the E. coli DsbA with the MdbA transmembrane domain rescued the pilus assembly defect at 30°C temperature (Fig. 5), thus providing additional support for conservation of the oxidative folding reaction mechanisms between Gram-positive and Gram-negative organisms.
As a vast number of C. diphtheriae exported proteins (~ 60%) secreted through the Sec translocon harbor two or more Cys residues, we hypothesized that MdbA may function as a general protein folding factor. To validate this conjecture, we probed the well-known exotoxin DT that contains two bona fide disulfide bonds. Production of full-length DT was significantly reduced in the ΔmdbA mutant with significant accumulation of degradation products (Fig. 4). By a combination of alkylation and furin-cleavage assays, we demonstrated that the mdbA mutant secretes a reduced form of DT, irrefutable evidence that MdbA is required for disulfide bond formation in vivo.
Because MdbA is required for optimal cell growth as well as the production of two virulence factors that are important for colonization and cytotoxicity, MdbA must be a major virulence factor for successful infection by corynebacteria. We tested this logical hypothesis with an animal survival assay using the guinea pig model of diphtheritic toxemia, expecting that the ΔmdbA mutant, similar to Δtox, would be nonvirulent. Indeed, while the majority of animals inoculated with wild-type C. diphtheriae succumbed to infection and died shortly, most of the animals exposed to the ΔmdbA mutant survived (Fig. 6). Notably, some animals infected with the ΔmdbA mutant were killed. This result seems somewhat surprising as C. diphtheriae ΔmdbA displays a growth defect at 37°C, the host’s normal body temperature. However, it is important to recall that DT production is normally triggered by unfavorable growth conditions, i.e. when cells are not dividing. An alkylation experiment did reveal that ΔmdbA secreted DT with reduced Cys residues, but a small portion of full-length toxin was not reactive (Fig. 5), suggesting that some of the toxin contained disulfide bonds. It is conceivable that spontaneous protein oxidation in the host environment permitted the proper folding of a sufficient amount of DT to cause the lethal infection in some, but not all of the animals. We propose that MdbA would serve as an ideal target to develop antimicrobials against Actinobacterial pathogens including M. tuberculosis. The fact that the ΔmdbA mutant is highly sensitive to β-lactams, a combination of MdbA inhibitors with additional antibiotics, may provide an effective antimicrobial strategy.
Finally, it is noteworthy to point out that the cell envelope of the genus Corynebacterium contains an outer layer of mycolic acids that may function as an outer membrane (Bayan et al., 2003; Burkovski, 2013). Indeed, freeze fracture cryo-EM provides physical evidence for the presence of this outer ‘mycomembrane’ in Mycobacterium bovis BCG, Mycobacterium smegmatis and C. glutamicum (Zuber et al., 2008). Thus, it is possible that C. diphtheriae possess an equivalent periplasmic compartment to regulate oxidative protein folding. We propose that as protein precursors emerge from the Sec translocon, they form a mixed-disulfide intermediate with MdbA (Fig. 7). Here, MdbA acts as a ‘place-holder’ (Kosuri et al., 2012) until a second Cys residue within the substrate is positioned to resolve the mixed-disulfide bond, which results in the formation of the disulfide linkage and the completion of substrate folding. In turn, the MdbA CxxC motif is reduced to an inactive state. It is likely that another thiol-oxidoreductase, we termed MdbB, would reoxidize MdbA. Its identity and functionality will be subject of future investigation.
Fig. 7.
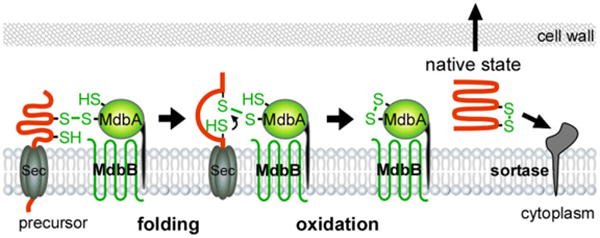
A model of MdbA-mediated posttranslocational protein folding. The transmembrane thiol-disulfide oxidoreductase MbdA enzyme forms a mixed disulfide bond with nascent precursors emerging from the Sec translocon, allowing the precursors to fold into a near-native state before MdbA-driven catalysis of disulfide bond formation, resulting in fully folded proteins for subsequent steps. An unidentified membrane-bound oxidoreductase termed MdbB is proposed to reoxidize MdbA.
Experimental procedures
Bacterial strains, primers, plasmids and media
Bacterial strains, primers and plasmids used in this study are listed in Tables S1 and S2 of Supplemental Information. Corynebacterium strains were grown in Heart Infusion (HI) broth or HI agar plates. E. coli DH5α and BL21, used for molecular cloning and protein purification, respectively, were grown in Luria Broth (LB) or agar. Kanamycin (Kan) was added at 50 μg ml−1 when needed.
Construction of recombinant plasmids
pMdbA
Primers mdbA_BamHl_F and mdbA_BamHl_R were designed to PCR-amplify the promoter and coding regions of C. diphtheriae mdbA with appending BamHI sites. The DNA fragment was digested with the appropriate restriction enzyme, and cloned into pCGL0243 pretreated with alkaline phosphatase and BamHI. The resulting plasmid was electroporated into NJ2.
pDsbATM
The primers dsbAEc_F and dsbAEc_R_BamHI were used to PCR amplify the extracellular regions of E. coli dsbA using Phusion DNA polymerase® (NEB) to generate blunt ends. The resulting product was 5′ phosphorylated and cut with BamHI. Segments encoding the promoter, RBS, and cytoplasmic and transmembrane domains for C. diphtheriae mdbA were amplified with primers PmdbA_F_BamHI and PmdbATM_R, and then digested with BamHI. Both DNA fragments then were ligated with pCGL0243 precut with BamHI and treated with alkaline phosphatase (NEB) to generate the recombinant plasmid, which was electroporated into NJ2.
Recombinant vectors using pMCSG7
To generate recombinant, His-tagged MdbA proteins, primers (see Tables S1 and S2) were designed to amplify the extracellular-coding regions of C. diphtheriae mdbA. The resulting PCR products were cloned into pMCSG7 using ligation-independent cloning (Stols et al., 2002). Purified DNA fragments were treated with LIC-competent T4 DNA polymerase (Novagen) and 2.5 μM dCTP. Meanwhile, pMCSG7, precut with SspI, was treated with LIC-competent T4 polymerase and dGTP to generate complementary overhangs between the linearized vector and mdbA. The products were then incubated over a gradient of temperatures (3 min at 70°C, 2 min at 65°C, 2 min at 60°C, 2 min at 55°C, 1 min at 50°C, 1 min at 45°C, 1 min at 40°C, 1 min at 35°C, 1 min at 30°C, 5 min at 25°C) to promote annealing. The resulting plasmids were used to transform E. coli DH5α and the insert was confirmed by DNA sequencing. The plasmids were then introduced into E. coli BL21 (DE3) for protein expression.
Site-directed mutagenesis of recombinant plasmids
To generate cysteine-to-alanine mutations within SpaA, inverse PCR was utilized using recombinant plasmids as templates (Table S2). Appropriate primers (Table S1) carrying the desired mutations were 5′ phosphorylated and used to PCR-amplify the plasmid templates with Phusion HF DNA polymerase (NEB). The resulting linear products were purified products, treated with ligase to reform the circular plasmids, and used to transform E. coli DH5α. DNA sequencing confirmed the desired mutations, and the plasmids were introduced to the appropriate strains (Table S2).
Generating gene deletions in C. diphtheriae
Nonpolar, in-frame deletion mutants were generated using the SacB counter-selection protocol as reported by Ton-That and Schneewind (2003). Briefly, approximately 1 Kb-fragments up- and downstream of the genes of interest were cloned into the integration plasmid pK19mobsacB expressing Kan resistance and sacB genes (Schafer et al., 1994). The resulting plasmid was introduced into E. coli S17-1 for conjugation with C. diphtheriae. Integration clones were selected for growth on Kan and nalidixic acid. To induce a double-crossover event leading to plasmid excision, C. diphtheriae were grown overnight without antibiotics. Loss of the integrated plasmid was selected for growth on HI agar plates containing 1% sucrose, which is toxic to cells expressing sacB. Gene deletions were identified by PCR and analyzed by Western blotting.
Cell fractionation and Western blotting
Overnight cultures of C. diphtheriae strains were used to inoculate fresh cultures (1:50 dilution). Cells grown at 30°C or 37°C to mid-log phase were normalized to an OD600 of 1.0, and subject to centrifugation to separate the medium (S) and cell fractions. C. diphtheriae were washed and re-suspended in hydrolase buffer [SMM, 0.5M sucrose, 10 mm MgCl2, phosphate buffered saline (PBS) pH 7.4], and then incubated with cell wall hydrolase enzymes at 37°C for 3 h. After treatment, the soluble cell wall fractions (W) were separated from the protoplasts (P) by centrifugation. P fractions were then washed in SMM buffer, resuspended in PBS containing 0.1% Triton-114, and subjected to three freeze/thaw cycles using dry ice-ethanol and 100°C water baths. When needed, the cytoplasmic fraction (C) was isolated by ultracentrifugation of lysed protoplasts prior to TCA precipitation. The S, W and C fractions were TCA precipitated and acetone washed. Protein samples were resuspended in reducing or nonreducing SDS loading buffer, heated at 60°C for 10 minutes, and separated on Tris-glycine gels. Proteins were detected with rabbit-raised antisera diluted in 5% milk (1:20 000 for α-SpaA; 1:20 000, α-MdbA; or 1:1000, α-DT) followed by horseradish peroxidase (HRP) (1:10 000) conjugated goat anti-rabbit IgG for detection by chemiluminescence.
Protein purification and crystallography
The recombinant MdbA protein for crystallography expressed from pMCSG7-MdbA was purified from E. coli cells grown in M9 medium supplemented with ampicillin (100 μg ml−1). A selenomethionine (SeMet) derivative of the expressed MdbA protein was prepared as previously described (Walsh et al., 1999). Harvested cells were disrupted by sonication, and clear lysates were obtained by centrifugation. The recombinant protein was purified by affinity chromatography using Ni-NTA (Qiagen) in the presence of 5 mM β-mercaptoethanol. After digestion with 0.15 mg TEV protease (ratio of 1.5:200) for 16 h at 4°C, the cleaved proteins were passed through a Ni-NTA column for removal of the TEV enzyme and cleaved N-terminal tags. Subsequently, the protein was further purified by gel-filtration on a HiLoad 16/60 Superdex 200 pg column (GE Healthcare) in 10 mM HEPES buffer pH 7.5, 200 mM NaCl and 1 mM DTT. The protein was concentrated up to 40 mg ml−1 concentration using Amicon Ultracel 10K centrifugal filters (Millipore).
The initial crystallization condition was determined with a sparse crystallization matrix at 4°C and 16°C temperatures using the sitting-drop vapor-diffusion technique with MCSG crystallization suite (Microlytic), Pi-minimal and Pi-PEG screens (Jena Bioscience) (Gorrec et al., 2011). Several conditions yielded diffraction quality crystals. The primary diffracting MdbA crystals were obtained from G9 conditions of Pi-PEG screen (2.9% PEG 350 MME, 34.3% PEG 600, 50 mM Tris buffer pH 7.6) at 16°C after 2 weeks. Due to poor quality and anisotropicity of diffraction, the crystals were used for microseeding using Seed Bead (Hampton Research) against all conditions of Pi-PEG screen at several temperatures. The best MdbA crystals were obtained from F10 conditions of Pi-PEG screen (4.3% PEG 2000, 28.6% PEG 550 MME, 50 mM Tris buffer pH 8.0) at room temperature. The MdbA crystals selected for data collection were washed in Parathone-N oil and flash-cooled in liquid nitrogen.
Single-wavelength X-ray diffraction data were collected at 100 K temperature at the 19-ID beamline of the Structural Biology Center (Rosenbaum et al., 2006) at the Advanced Photon Source at Argonne National Laboratory using the program SBCcollect. The intensities were integrated and scaled with the HKL3000 suite (Minor et al., 2006).
The structure was determined by single-wavelength anomalous dispersion (SAD) phasing using HKL3000 suite (Adams et al., 2002) incorporating SHELXC, SHELXD, SHELXE (Sheldrick, 2010), MLPHARE, SOLVE/RESOLVE (Terwilliger, 2003) programs. Several rounds of manual adjustments of structure models using COOT (Emsley and Cowtan, 2004) and refinements with Refmac program (Murshudov et al., 1997) from CCP4 suite (Collaborative Computational Project, 1994) were done. The stereochemistry of the structure was validated with PHENIX suite (Adams et al., 2002) incorporating MOLPROBITY (Davis et al., 2004) tools. A summary of data collection and refinement statistics is given in Tables 1 and 2. The MdbA atomic coordinates and structure factors were deposited into the Protein Data Bank as 5C00.
Electron microcopy
Bacteria grown on HIA plates or liquid media were suspended and washed in 0.1M NaCl, and then resuspended in PBS. To examine the cells by electron microscopy, 7 μL of culture was placed onto carbon-coated nickel grids (Electron Microscopy Sciences) for 1 min, washed three times with sterile water and then stained with 1% uranyl-acetate for another minute. For immuno-gold labeling, 7 μL of culture was placed onto the grids for 1 min and then washed in PBS 1% BSA three times. The grids were subsequently blocked in PBS 1% BSA with 0.1% gelatin for 1 h at room temperature. Pili were stained with undiluted depleted α-SpaA diluted in PBS 1% BSA for 1 h, and then washed three times in PBS 1% BSA. Grids were blocked again in gelatin for 30 min, washed once and then stained with secondary antibody conjugated to 12 nm gold particles diluted 1:20 in PBS 1% BSA for 1 h. Finally, the samples were washed five times with sterile water and stained with 1% uranyl acetate for 1 min. All samples were viewed using a JEOL JEM-1400 electron microscope.
Van-FL fluorescence microscopy
Overnight cultures of C. diphtheriae were diluted 1:50 and then grown at 30°C or 37°C until reaching log phase. A 1:1 mixture of vancomycin (Van) and Van-FL was added to cultures at a concentration of 1 μg ml−1, and incubated for 10 min with agitation at 37°C. The samples were then placed on agar pads and viewed by a fluorescence microscope using excitation/emission wavelengths 504 nm/510 nm.
Spot dilution assays
Spot dilutions of C. diphtheriae were performed using overnight cultures grown at 30°C. To test for temperature sensitivity, C. diphtheriae cultures were spotted (10−3–10−6) onto HI agar plates, and then incubated at 37°C for 24 h or 30°C for 48 h. To test for antibiotic sensitivity, C. diphtheriae were spotted (10−3–10−6) into HI agar plates containing various concentrations of ampicillin or penicillin, and incubated at 30°C for 48 h. The concentrations of antibiotics chosen were based on previous calculations of antimicrobial MICs for C. diphtheriae (Soriano et al., 1995).
Alkylation of pilus proteins and diphtheria toxin
C. diphtheriae SpaA monomers were collected from the medium fraction of C. diphtheriae HT3, a mutant that secretes monomeric SpaA pilins into the culture medium due to the absence of sortases (Swaminathan et al., 2007). Bacteria were grown to mid-log phase, SpaA were isolated from the medium fraction of mid-log phase grown bacteria by centrifugation, TCA precipitated and acetone washed. To alkylate SpaA, samples were reduced in DTT-containing buffer (100 mM Tris-HCl, 1% SDS, 100 mM DTT, pH 8) at room temperature for 1 h, followed by TCA precipitation and acetone wash to remove the DTT. The resulting pellets were treated with Mal-PEG in alkylation buffer (100 mM Tris-HCl pH 6.8, 1% SDS, 20 mM Mal-PEG) at room temperature for 1 h, followed by TCA precipitation and acetone wash. Protein samples were then dissolved in SDS-loading buffer and separated by 3–20% Tris-glycine gels for immunoblotting with α-SpaA.
To alkylate DT, overnight cultures of C. diphtheriae were diluted 1:50 and grown at 30°C until reaching an OD600 between 0.2 and 0.3. At this point, 10 mgmL−1 of iron chelator ethylenediamine-di-(o-hydroxyphenylacetic) acid (EDDA) was added to the cell cultures to induce DT production for 2 h. DT was isolated from the culture medium by centrifugation, and then TCA precipitated and acetone washed. Alkylation of DT by Mal-PEG was then performed as described in the previous section. DT protein was detected by immunoblotting with a monoclonal antibody generated against the A fragment of DT (Santa Cruz Biotechnology).
Furin cleavage of diphtheria toxin
Similar to the DT alkylation experiment, EDDA was added to liquid cultures of C. diphtheriae to induce toxin production. DT was collected from the culture medium by centrifugation, and then concentrated using an Ultracel-30K centrifugal filter (Amicon). Equal volumes of sample were then resuspended in Furin cleavage buffer (100 mM HEPES pH 7.0, 1 mM Cacl2), combined with 2U of Furin convertase (Thermo-Scientific) and incubated at 37°C for 2h. The reactions were then terminated by the addition of SDS loading buffer with or without 2-β-mercaptoethanol, and DT was detected using a monoclonal antibody generated against the A fragment of DT (Santa Cruz Biotechnology).
Infection of guinea pigs
For infection, mid-log phase C. diphtheriae cells grown in HI broth at 30°C were collected by centrifugation, washed and resuspended in PBS. Groups of six Hartley guinea pigs (4–5 week-old) were infected via intraperitonal injections with 2.5 × 107 CFUs of each bacterial strain. Animal survival was examined over 7 days, but severely moribund guinea pigs were humanely euthanized. The survival curves were analyzed using Mantel–Cox and chi-square tests.
The procedure above was approved by the Institutional Animal Care and Use Committee (IACUC) for the University of Texas Health Science Center at Houston (UTH). The UTH IACUC adheres to the NIH Office of Laboratory Animal Welfare standards.
Supplementary Material
Acknowledgments
We thank Timothy Fothergill and Shilpa Muralidhar for technical assistance, members of the Structural Biology Center at Argonne National Laboratory for their help in conducting X-ray diffraction data collection, and our laboratory members for the critical review and discussion of the manuscript. M.E.R.-R. was previously supported by the Predoctoral Training Program in Molecular Basis of Infectious Diseases, National Institute of Allergy and Infectious Diseases (NIH Grant T32 AI55449). This work was supported by the National Institute of General Medical Sciences grant GM094585 (to J.O. and A.J.) and the National Institute of Dental and Craniofacial Research under award numbers F31DE024004 (to M.E.R.-R.) and DE025015 (to H.T.-T.). Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayan N, Houssin C, Chami M, Leblon G. Mycomembrane and S-layer: two important structures of Corynebacterium glutamicum cell envelope with promising biotechnology applications. J Biotechnol. 2003;104:55–67. doi: 10.1016/s0168-1656(03)00163-9. [DOI] [PubMed] [Google Scholar]
- Boquet P, Pappenheimer AM., Jr Interaction of diphtheria toxin with mammalian cell membranes. J Biol Chem. 1976;251:5770–5778. [PubMed] [Google Scholar]
- Burkovski A. Cell envelope of corynebacteria: structure and influence on pathogenicity. ISRN Microbiol. 2013;2013:935736. doi: 10.1155/2013/935736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim N, Harmston CA, Guzman DJ, Goulding CW. Structural and biochemical characterization of the essential DsbA-like disulfide bond forming protein from Mycobacterium tuberculosis. BMC Struct Biol. 2013;13:23. doi: 10.1186/1472-6807-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Collier RJ. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon. 2001;39:1793–1803. doi: 10.1016/s0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- Crow A, Lewin A, Hecht O, Carlsson Moller M, Moore GR, Hederstedt L, Le Brun NE. Crystal structure and biophysical properties of Bacillus subtilis BdbD. An oxidizing thiol:disulfide oxidoreductase containing a novel metal site. J Biol Chem. 2009;284:23719–23733. doi: 10.1074/jbc.M109.005785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Mellroth P, Bernsel A, Neiers F, Normark S, von Heijne G, Henriques-Normark B. Disulfide bond formation and cysteine exclusion in gram-positive bacteria. J Biol Chem. 2010;285:3300–3309. doi: 10.1074/jbc.M109.081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey L, Ng CK, Halperin SA, Lee SF. Functional analysis of paralogous thiol-disulfide oxidore-ductases in Streptococcus gordonii. J Biol Chem. 2013;288:16416–16429. doi: 10.1074/jbc.M113.464578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorenbos R, Stein T, Kabel J, Bruand C, Bolhuis A, Bron S, et al. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J Biol Chem. 2002;277:16682–16688. doi: 10.1074/jbc.M201158200. [DOI] [PubMed] [Google Scholar]
- Dumoulin A, Grauschopf U, Bischoff M, Thony-Meyer L, Berger-Bachi B. Staphylococcus aureus DsbA is a membrane-bound lipoprotein with thiol-disulfide oxidoreductase activity. Arch Microbiol. 2005;184:117–128. doi: 10.1007/s00203-005-0024-1. [DOI] [PubMed] [Google Scholar]
- Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gorrec F, Palmer CM, Lebon G, Warne T. Pi sampling: a methodical and flexible approach to initial macromolecular crystallization screening. Acta Crystallogr D Biol Crystallogr. 2011;67:463–470. doi: 10.1107/S0907444911008754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras B, Kurz M, Jarrott R, Shouldice SR, Frei P, Robin G, et al. Staphylococcus aureus DsbA does not have a destabilizing disulfide. A new paradigm for bacterial oxidative folding. J Biol Chem. 2008;283:4261–4271. doi: 10.1074/jbc.M707838200. [DOI] [PubMed] [Google Scholar]
- Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, Martin JL. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138:1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci USA. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ito K. Respiratory chain strongly oxidizes the CXXC motif of DsbB in the Escherichia coli disulfide bond formation pathway. EMBO J. 1999;18:1192–1198. doi: 10.1093/emboj/18.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooi-Pol MM, Reilman E, Sibbald MJ, Veenstra-Kyuchukova YK, Kouwen TR, Buist G, van Dijl JM. Requirement of signal peptidase ComC and thiol-disulfide oxidoreductase DsbA for optimal cell surface display of pseudopilin ComGC in Staphylococcus aureus. Appl Environ Microbiol. 2012;78:7124–7127. doi: 10.1128/AEM.01565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri P, Alegre-Cebollada J, Feng J, Kaplan A, Ingles-Prieto A, Badilla CL, et al. Protein folding drives disulfide formation. Cell. 2012;151:794–806. doi: 10.1016/j.cell.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London E. Diphtheria toxin: membrane interaction and membrane translocation. Biochim Biophys Acta. 1992;1113:25–51. doi: 10.1016/0304-4157(92)90033-7. [DOI] [PubMed] [Google Scholar]
- Makmura L, Hamann M, Areopagita A, Furuta S, Munoz A, Momand J. Development of a sensitive assay to detect reversibly oxidized protein cysteine sulfhydryl groups. Antioxid Redox Signal. 2001;3:1105–1118. doi: 10.1089/152308601317203611. [DOI] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, Bardwell JC, Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- Meima R, Eschevins C, Fillinger S, Bolhuis A, Hamoen LW, Dorenbos R, et al. The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J Biol Chem. 2002;277:6994–7001. doi: 10.1074/jbc.M111380200. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution – from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Pappenheimer AM, Jr, Uhr JW, Yoneda M. Delayed hypersensitivity. I. Induction of hypersensitivity to diphtheria toxin in guinea pigs by infection with Corynebacterium diphtheriae. J Exp Med. 1957;105:1–9. doi: 10.1084/jem.105.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L, Heras B, Duprez W, Walden P, Halili M, Kurth F, et al. essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases. Acta Crystallogr D Biol Crystallogr. 2969c;69:1981–1994. doi: 10.1107/S0907444913017800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon ME, Ton-That H. Assembly and function of Corynebacterium diphtheriae pili. In: Burkovski A, editor. Corynebacterium Diphtheriae and Related Toxigenic Species. Netherlands: Springer; 2013. p. 7. [Google Scholar]
- Reardon-Robinson ME, Osipiuk J, Chang C, Wu C, Jooya N, Joachimiak A, et al. A disulfide bond-forming machine is linked to the sortase-mediated pilus assembly pathway in the gram-positive bacterium Actinomyces oris. J Biol Chem. 2015;290:21393–21405. doi: 10.1074/jbc.M115.672253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Stephan D, Xu Z, Zheng Y, Tang D, Harrison RS, et al. Properties of the thioredoxin fold super-family are modulated by a single amino acid residue. J Biol Chem. 2009;284:10150–10159. doi: 10.1074/jbc.M809509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Das A, Ton-That H. Adhesion by pathogenic corynebacteria. Adv Exp Med Biol. 2011;715:91–103. doi: 10.1007/978-94-007-0940-9_6. [DOI] [PubMed] [Google Scholar]
- Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, et al. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synchrotron Radiat. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouldice SR, Heras B, Walden PM, Totsika M, Schembri MA, Martin JL. Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid Redox Signal. 2011;14:1729–1760. doi: 10.1089/ars.2010.3344. [DOI] [PubMed] [Google Scholar]
- Soriano F, Zapardiel J, Nieto E. Antimicrobial susceptibilities of Corynebacterium species and other non-spore-forming gram-positive bacilli to 18 antimicrobial agents. Antimicrob Agents Chemother. 1995;39:208–214. doi: 10.1128/aac.39.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stols L, Gu M, Diechkman L, Raffer R, Collart FL, Donnelly ML. A new vector for high-throughput ligation independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Um SH, Kim JS, Lee K, Ha NC. Structure of a DsbF homologue from Corynebacterium diphtheriae. Acta Crystallogr F Struct Biol Commun. 2014;70:1167–1172. doi: 10.1107/S2053230X14016355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda A, Amako K. Growth of the surface of Corynebacterium diphtheriae. Microbiol Immunol. 1983;27:663–671. doi: 10.1111/j.1348-0421.1983.tb00629.x. [DOI] [PubMed] [Google Scholar]
- Valbuena N, Letek M, Ordonez E, Ayala J, Daniel RA, Gil JA, Mateos LM. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol Microbiol. 2007;66:643–657. doi: 10.1111/j.1365-2958.2007.05943.x. [DOI] [PubMed] [Google Scholar]
- Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: ultrafast protein structure determination. Acta Crystallogr D Biol Crystallogr. 1999;55:1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- Young RM, Mood GM. Effect of penicillin on infection of guinea pigs with Corynebacterium diphtheriae. J Bacteriol. 1945;50:205–212. doi: 10.1128/JB.50.2.205-212.1945. [DOI] [PubMed] [Google Scholar]
- Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


