Abstract
Defects in cilia cause a spectrum of diseases known as ciliopathies. Nephronophthisis, a ciliopathy, is the most common genetic cause of renal disease. Here, I cloned and analysed a nephronophthisis-related gene ift-139 in Caenorhabditis elegans. ift-139 was exclusively expressed in ciliated neurons in C. elegans. Genetic and cellular analyses suggest that ift-139 plays a role in retrograde intraflagellar transport and is required for cilia formation. A homologous point mutation that causes ciliopathy disrupted the function of ift-139 in C. elegans. ift-139 is an orthologue of human TTC21B, mutations in which are known to cause nephronophthisis 12 and short-rib thoracic dysplasia 4. These results suggest that ift-139 is evolutionarily conserved and fundamental to the formation of cilia.
Cilia and flagella are essential for cellular motility, sensation of extracellular stimuli and development of multicellular animals1,2. Motile cilia generate fluid flow and cell motility, while non-motile cilia work as receptors for various chemical and mechanical signalling. Though their physiological roles are different, the formation of both motile and non-motile cilia depends on a common molecular mechanism called intraflagellar transport (IFT)1. The IFT mechanism is very well conserved among unicellular ciliates and higher eukaryotes1,3. Disruption of IFT leads to ciliopathies including retinal degeneration, Bardet-Biedl syndrome, polycystic kidney disease, nephronophthisis and defects in the establishment of organ laterality (situs inversus)1,4,5,6,7,8. The cilia cytoskeleton is composed of microtubules, microtubule-associated proteins and dyneins9. The microtubule-dependent motors, kinesin-2 and cytoplasmic dynein-2, bind to the IFT-A and IFT-B multiprotein complexes to carry out IFT. IFT-A and IFT-B complexes are involved in the retrograde and anterograde transport of components of cilia, respectively. The molecular mechanism of IFT was first established in a unicellular ciliate Chlamydomonas reinharditii, where a combination of biochemistry and genetics revealed that Chlamydomonas IFT complex contain at least 16 proteins10,11.
Caenorhabditis elegans (C. elegans) is a good model for investigating the cell biology of ciliopathies3,12,13. Most genes associated with ciliopathies, including genes encoding IFT complex proteins, are conserved in C. elegans and mutations in some of these genes have been shown to lead to defects in cilia morphology and function in C. elegans13,14,15,16,17,18. For instance, che-2 and dyf-2, originally identified as a chemotaxis mutant gene and dye-filling mutant gene, respectively encode homologues of the IFT80 and IFT144 subunits of the IFT complex19,20. Mutations in IFT80 and IFT144 genes are genetic causes of human ciliopathy21,22 and C. elegans che-2 and dyf-2 mutants have behavioural defects because of degenerated cilia.
Mutations in the TTC21B gene, which encodes the human IFT139 subunit, are causes of human ciliopathies such as NPHP12 and short-rib thoracic dysplasia 423,24. C. elegans has proven to be a valuable model for the study of the function of many cilia proteins and ciliopathies. The presence of IFT139 subunit is predicted by C. elegans genomic analysis25. A promoter analysis has shown that a candidate IFT139 homologue, ZK328.7, is expressed in ciliated sensory neurons26. However, the function and the protein localization of C. elegans orthologue of TTC21B/IFT139 has not been analyzed in detail to date. Here, I cloned and analyzed the nematode ift-139/IFT139. Moreover the effect of a mutation in ift-139, resulting in ciliopathy, is reported.
Results
ZK328.7 is a cilia-related gene
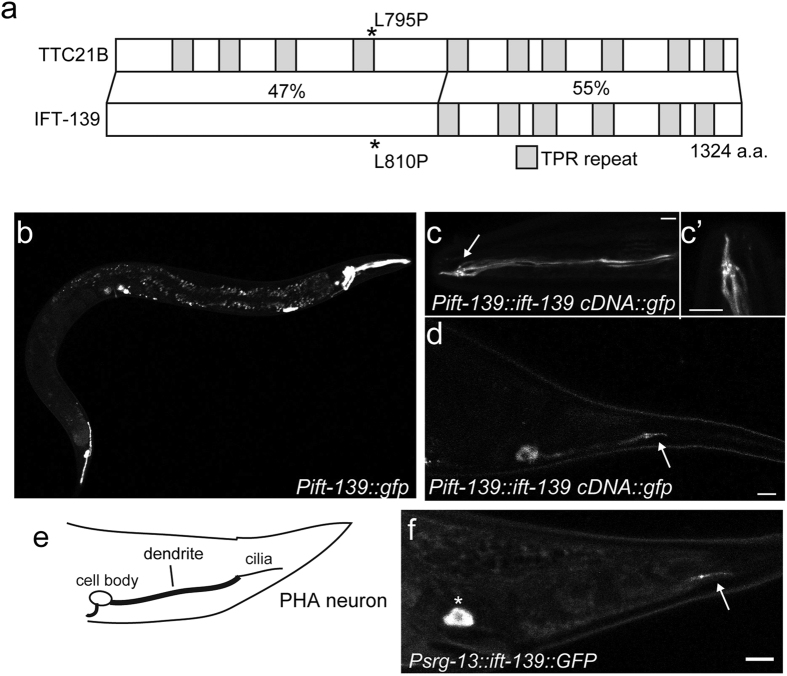
To investigate the molecular mechanism of ciliopathy, C. elegans genes orthologous to human ciliopathy genes were determined. ZK328.7 is an orthologue of the human TTC21B gene, mutations that cause congenital diseases such as NPHP12 and short-rib thoracic dysplasia 4 (Fig. 1a). TTC21B encodes the IFT139 subunit in humans27. IFT139 was originally identified as a component of the IFT complex in Chlamydomonas10,28. Because of the homology of the protein encoded by ZK328.7 with other IFT139 protein family members, the gene was renamed ift-139. Based on the predicted sequence, I performed polymerase chain reaction (PCR) from wild-type cDNA and cloned ift-139. While it is predicted that ift-139 encodes two isoforms, the longer isoform which encodes a 1324-amino acid protein was obtained by 5′ UTR and 3′ UTR primer pairs. IFT-139 has at least six TPR domains, as revealed with the SMART algorithm29. The feature is similar to other IFT139 family members.
Figure 1. Cloning and characterisation of ift-139.
(a) Comparison between TTC21B and IFT-139. Domains are predicted with the SMART algorithm. Both TTC21B and IFT-139 contain multiple TPR domains. The C-terminal halves of both proteins are predicted to have six TRP domains and the identity is 55%. While the N-terminal half of IFT-139 is not predicted to contain TPR repeats, the identity of the N-terminal half was 47%. (b) Expression pattern of the GFP signal under the ift-139 promoter. Bar, 100 μm. (c,d) Localisation of IFT-139::GFP expressed under the ift-139 promoter. Head neurons (c), amphid cilia (c’) and phasmid neurons (d) are illuminated. (c’) is a zoom of the cilia region in (c). Arrows indicate cilia. Bars, 5 μm. (e,f) Schematic of the PHA neuron revealed by the srg-13 promoter (e) and the IFT-139::GFP signal in the PHA neuron (f). The asterisk and arrow indicate cell body and cilia, respectively. Bar, 5 μm.
IFT-139 is expressed in ciliated neurons and localises to basal bodies and cilia
To assess the cellular expression of ift-139, an 800-bp fragment upstream of the start codon, spanning the putative promoter region, was cloned into a Green Fluorescent Protein (GFP) expression vector and injected into wild-type nematodes. Transgenic animals were established and observed under the fluorescent microscope. GFP expression was observed in ciliated sensory neurons in both head and tail regions (Fig. 1b).
Next, the open reading frame encoding ift-139 (1324 amino acids) was fused with GFP and expressed from the promoter described above (Fig. 1c,d). IFT-139::GFP was observed in ciliated sensory neurons. In the amphid neurons, the GFP signal was diffuse in dendrites and concentrated to the base of cilia (Fig. 1c,c’). Moreover, the cilia signal was stronger than dendrite. In phasmid PHA and PHB neurons, most of the GFP signals were observed at the base of cilia and in the cilia (Fig. 1d). Phasmid dendritic signals were relatively weak compared to amphid neurons. To observe the localisation of IFT-139 at single-cell resolution, IFT-139::GFP was expressed from the PHA neuron-specific promoter, srg-13 promoter (Fig. 1e). Once again, the IFT-139::GFP signal was localised to the base of cilia and in the cilia of PHA neuron (Fig. 1f). These data suggest that IFT-139 may play a role in cilia.
ift-139 is required for the formation and function of cilia in C. elegans
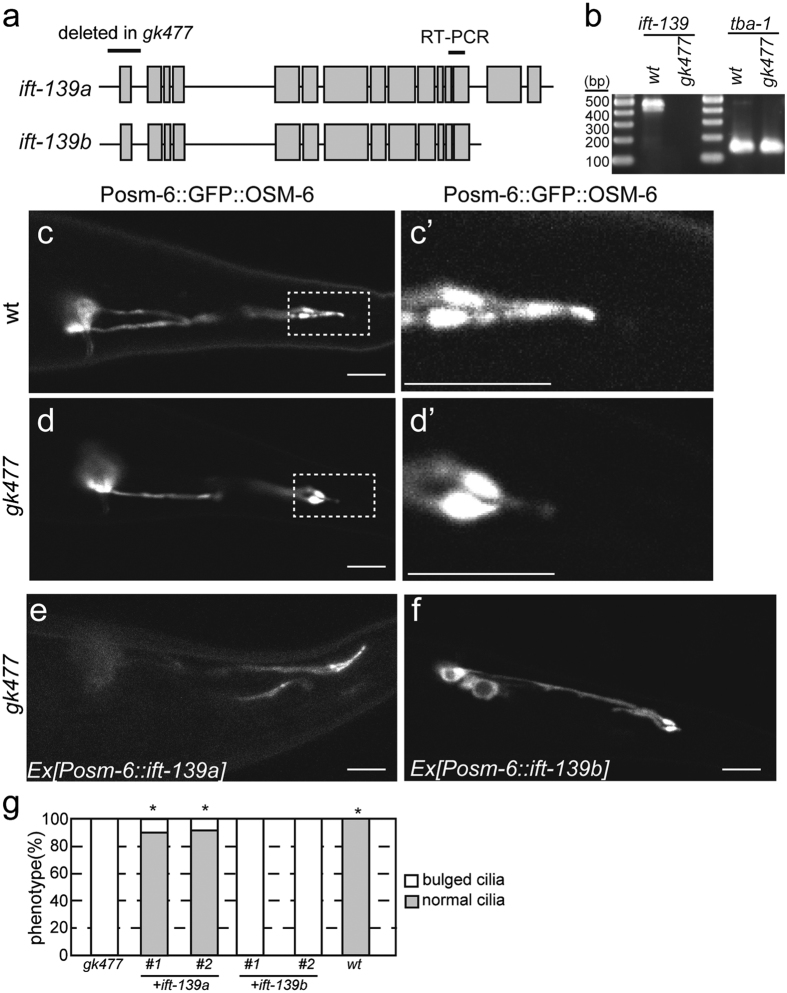
I searched WormBase (www.wormbase.org) and found that gk477 is a deletion mutant of ift-139 in which the first exon of the gene is deleted (Fig. 2a). Though it is predicted that the ift-139 genome encodes two isoforms, both isoforms share the promoter and the first exon, suggesting that both isoforms are deleted in gk477. To test whether gk477 affects the expression of ift-139, reverse transcription polymerase chain reaction (RT-PCR) was performed using primers that can detect both isoforms (Fig. 1a). Results of RT-PCR showed that the expression of ift-139 was under the detectable level in gk477 (Fig. 2b), suggesting that gk477 is a null mutant.
Figure 2. Analysis of the ift-139 mutant.
(a) Schematic of the ZK328.1/ift-139 genomic region. Two isoforms are predicted in the database. The first exon of both isoforms is deleted in gk477. The region amplified in panel (b) is indicated. (b) RT-PCR was performed using primer pairs that can amplify ift-139 and tba-1. Note that ift-139 is not detected in gk477. (c–g) mnIs17[osm-6p::osm-6::gfp] is crossed with wt and ift-139(gk477) worms and the GFP signal in PHA and PHB neurons was observed. Representative images showing the morphology of wild-type phasmid cilia (c,c’), ift-139(gk477) phasmid cilia (d,d’) and ift-139(gk477) expressing ift-139a cDNA (e) and ift-139b cDNA (f). (c’,d’) shows the boxed area in (c,d). (g) Graph showing the results of the rescue experiments. *p < 0.01, compared with ift-139. Chi-square test with Bonferroni correction. n = 40 worms.
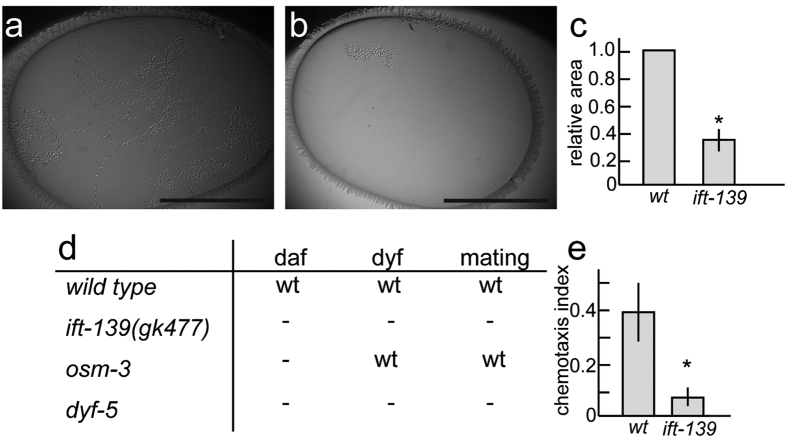
Cilia morphology was investigated in nematodes carrying the ift-139(gk477) mutation using the well-established and widely used cilia marker, mnIs1730. This marker stably expresses OSM-6::GFP in ciliated neurons under the osm-6 promoter. It was found that nematodes carrying the ift-139(gk477) mutation had abnormal cilia whose length were significantly shorter than wild type (Fig. 2c,d; wt: 6.4 ± 0.7 μm; ift-139(gk477): 3.9 ± 0.7 μm; mean ± standard deviation; n = 27 cilia; p < 0.01, t-test). Cilia were also more bulged in ift-139(gk477) than in wild-type animals (Fig. 2d,d’). To test that this phenotype was caused by the ift-139(gk477) mutation, and to determine whether the effect was cell autonomous, wild-type ift-139 cDNA was expressed specifically in ciliated neurons of the ift-139(gk477) mutant from the osm-6 promoter. The activity of both long and short isoforms of ift-139 was tested. As a result, the cilia morphology defect was rescued by expressing ift-139a (long isoform) but not ift-139b (short isoform) (Fig. 2e,f). Thus, I focused on the long isoform in this manuscript in the following experiments. To confirm that the morphological defect revealed by the GFP marker reflects abnormalities in vivo, phenotypes that are induced by ciliary defects were investigated. First, behaviour of ift-139(gk477) was observed. Single worms were placed on OP50 feeder bacteria and incubated overnight. While wild-type worms moved all over the feeder bacteria, ift-139(gk477) movement was restricted to a narrow area (Fig. 3a,b, 38% compared with wt). Nevertheless, unlike unc mutants, the speed of worm motility was not changed significantly when they were moving on the agarose plate. The phenotype is similar to those observed in ift-74 and ift-81 mutants, in which cilia are disrupted31. Next, dauer formation, dye-filling and mating behaviours were examined; these are known to require normal cilia function and morphology32,33,34. Consistent with the abnormal cilia morphology, defects in all of these processes were observed in ift-139(gk477) nematodes (Fig. 3d). In the ift-139-expressing strain (Fig. 2e), normal mating behaviour was recovered. Furthermore, previous studies have shown that C. elegans is attracted to diacetyl, a volatile substance, and that cilia are required for this chemotaxis35. We therefore calculated the chemotaxis index to diacetyl as described, comparing wild-type and ift-139(gk477) mutants36. As expected, chemotaxis was impaired, with the chemotaxis index of the ift-139(gk477) mutant significantly lower than wild type (Fig. 3e). Collectively, these phenotypes indicate that cilia morphology and function require ift-139.
Figure 3. Gross phenotype of ift-139 mutant.
(a–c) A single worm was placed on OP50 feeder bacteria and incubated overnight. (a,b) Representative image of wild type (a) and ift-139(b). (c) Relative moving area was calculated. Mean ± standard deviation (S.D.). *p < 0.01, t-test, n = 10. Note that ift-139(gk477) mutants move in a narrow area compared with wild type. Bars, 1 cm. (d) Gross phenotypes in ift-139 mutants were compared with wild type, osm-3, and dyf-5 mutants. daf, dauer formation, dyf, dye filling and mating: male mating. wt means the phenotype was comparable to wt, - means the defects in the phenotype. (e) Chemotaxis to diacetyl was observed and chemotaxis index was calculated as described in the materials and methods. Mean ± S.D. n = 10 experiments. *p < 0.01, t-test.
Human TTC21B can partially rescue ift-139(gk477)
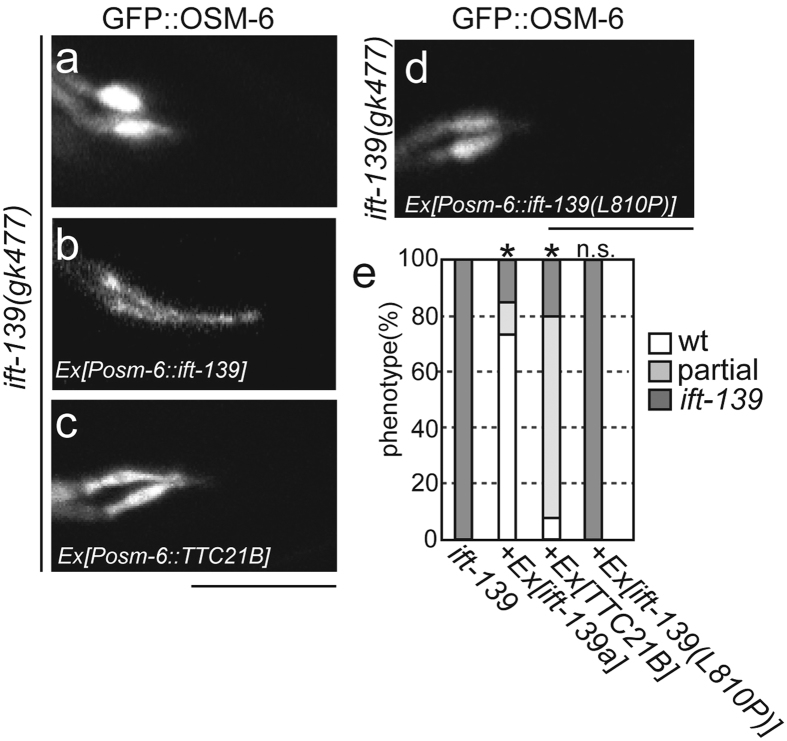
ift-139 is the nematode orthologue of TTC21B. To test whether the function of ift-139 is evolutionarily conserved, the human TTC21B gene was cloned and fused with the osm-6 promoter. Then, the vector was injected into ift-139(gk477) mutants. As a result, expressing human TTC21B could partially rescue cilia morphology; cilia length was longer in ift-139(gk477) mutants in the presence of human IFT139 than without, but shorter than wild-type cilia (Fig. 4a–c). The rescued cilia was more wild-type like morphology, but still more bulged than wild type. Statistical analysis supports the observations (Fig. 4e).
Figure 4. ift-139 is related to the human TTC21B gene.
ift-139(gk477); mnIs17[osm6p::osm-6::gfp] (a) was rescued with the expression of nematode ift-139 cDNA (b), human TTC21B (c) and nematode ift-139 with the L810P mutation (d). Representative photographs are shown. Bars, 5 μm. (e) Graph showing the results of the rescue experiments. *p < 0.01, n.s, p > 0.05, compared with ift-139. Chi-square test with the Bonferroni correction. n = 40 worms.
A ciliopathy mutation disrupts the function of ift-139
It has been shown that TTC21B mutations cause autosomal recessive ciliopathies such as NPHP12 and short-rib thoracic dysplasia 423,24,27. Most TTC21B mutations causing ciliopathies are deletions27, although some point mutations have been described, including a point mutation leading to an L795P substitution in human TTC21B. The L795 residue is conserved in C. elegans IFT-139 (L810). We postulated that a similar substitution in this conserved amino acid would result in ciliary defects in C. elegans. Therefore, we introduced a transgene encoding ift-139 with the corresponding mutation, L810P, into C. elegans ift-139(gk477) and assessed its effect on the activity of nematode IFT-139. I tested the long isoform of ift-139 in this experiment. While expression of wild-type ift-139 could recover the cilia morphology of ift-139(gk477) mutants, expression of ift-139(L810P) could not (Fig. 4b,d,e). The morphology of cilia was still bulb like and could not be discriminated from ift-139(gk477) in IFT-139(L810P)-expressing worms. These rescue experiments by human TTC21B and IFT-139(L810P) suggest that the structure and function are well, but not perfectly, conserved between human TTC21B and nematode IFT-139.
Genetic interaction suggests IFT-139 plays a role in retrograde intraflagellar transport
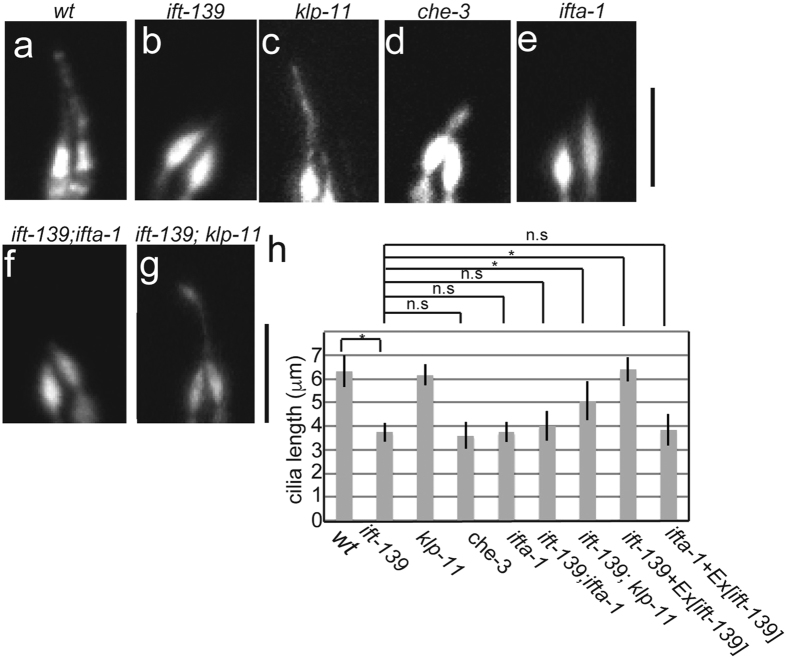
Kinesin-2 and cytoplasmic dynein-2 anterogradely and retrogradely transport IFT particles in cilia. There are two types of kinesin-2: a heterotetramer composed of KLP-11, KLP-20 and KAP-1 and a homodimer composed of OSM-337,38. Cytoplasmic dynein-2 is encoded by che-339. To determine whether ift-139 plays a role in anterograde or retrograde transport, cilia morphology was genetically analysed (Fig. 5a–h). By comparing worms carrying mutations in ift-139 with those carrying mutations in klp-11, affecting anterograde transport, and che-3, affecting retrograde transport, it was found that ift-139 mutations phenocopied che-3 but not klp-11 mutations (Fig. 5a–d). In klp-11 mutants, while the OSM-6::GFP signal was dimmer than wild type, the length of PHA/PHB cilia was not changed significantly (Fig. 5a,c; wt: 6.3 ± 0.67 μm; klp-11: 6.2 ± 0.45 μm; mean ± S.D.). The phenotype was, however, clearly different from ift-139 mutants that have shorter and bulb-like phasmid cilia (Fig. 5b; ift-139: 3.7 ± 0.40 μm; mean ± S.D.). In contrast, che-3 had shorter and bulged phasmid cilia as has been previously described (Fig. 5d; che-3: 3.6 ± 0.55 μm; mean ± S.D.)39. These genetic data suggested that IFT-139 is involved in retrograde IFT. The IFT-A complex is known to contribute to retrograde IFT40. One component of this complex is IFT122, encoded by ifta-141. To further confirm that ift-139 is involved in retrograde transport, the phenotypes of ift-139 and ifta-1 mutants were compared. The ifta-1 mutant had shorter and bulb-like cilia, similar to ift-139 (Fig. 5e; ifta-1: 3.8 ± 0.43 μm; mean ± S.D.). Next, a double mutant was generated. Cilia morphology of the ift-139; ifta-1 double mutant could not be discriminated from ift-139 and ifta-1 single mutants (Fig. 5b,e,f). The cilia length of ift-139; ifta-1 was 4.0 ± 0.62 μm (mean ± S.D.), that is statistically identical to the ift-139 single mutant. In contrast, an ift-139; klp-11 double mutant had longer cilia than the ift-139 single mutant (Fig. 5g,h; 6.4 ± 0.51 μm; mean ± S.D). In this double mutant, cilia morphology was still abnormal but more similar to wild-type rather than ift-139 mutants (Fig. 5g). These results again suggested that ift-139 is a component of the IFT-A complex and contributes to retrograde IFT.
Figure 5. Genetic analysis of ift-139.
Cilia morphology was observed in indicated mutants using the mnIs17 marker. Representative images of wt (a), ift-139 (b), klp-11 (c), che-3 (d), ifta-1 (e), ift-139; ifta-1 (f), ift-139; klp-11 (g). Bars, 5 μm. (h) Cilia length was measured and shown as a bar graph. *p < 0.05, n.s., P > 0.05. One-way analysis of variance. n = 28.
IFT-139 movement in cilia
The motility of IFT-139 was observed in phasmid cilia. The long isoform of IFT-139 was expressed as a GFP fusion as described above (Fig. 1c,d) and the signal was observed by spinning disk confocal microscopy. Many IFT-139::GFP-positive particles were bidirectionally moving in the cilia (Fig. 6a,b and Supplemental Movie S1). The speed of anterograde particles was measured at 0.72 ± 0.11 μm/s and 1.31 ± 0.11 μm/s in middle and distal segments, respectively (Fig. 6a–c). The speed of retrograde transport was 1.21 ± 0.32 μm/s and 1.11 ± 0.31 μm/s in middle and distal segments, respectively (Fig. 6d). These biphasic and monophasic transport parameters in anterograde and retrograde transport are almost identical to those previously found for the IFT-particle::GFP37,38,41. To analyse the effect of disease-associated mutations on the motility of IFT-139, IFT-139(L810P) was fused with GFP and introduced into wild-type worms. Interestingly, in both anterograde and retrograde transport, the motility of IFT-139(L810P) was comparable to that of wild-type IFT-139 in wild-type worms (Fig. 6c,d). However, the mutant protein could not rescue ift-139(gk477), suggesting that IFT-139(L810P) is normally incorporated into the IFT complex but is not functional in vivo.
Figure 6. Motility of IFT139 in phasmid cilia.
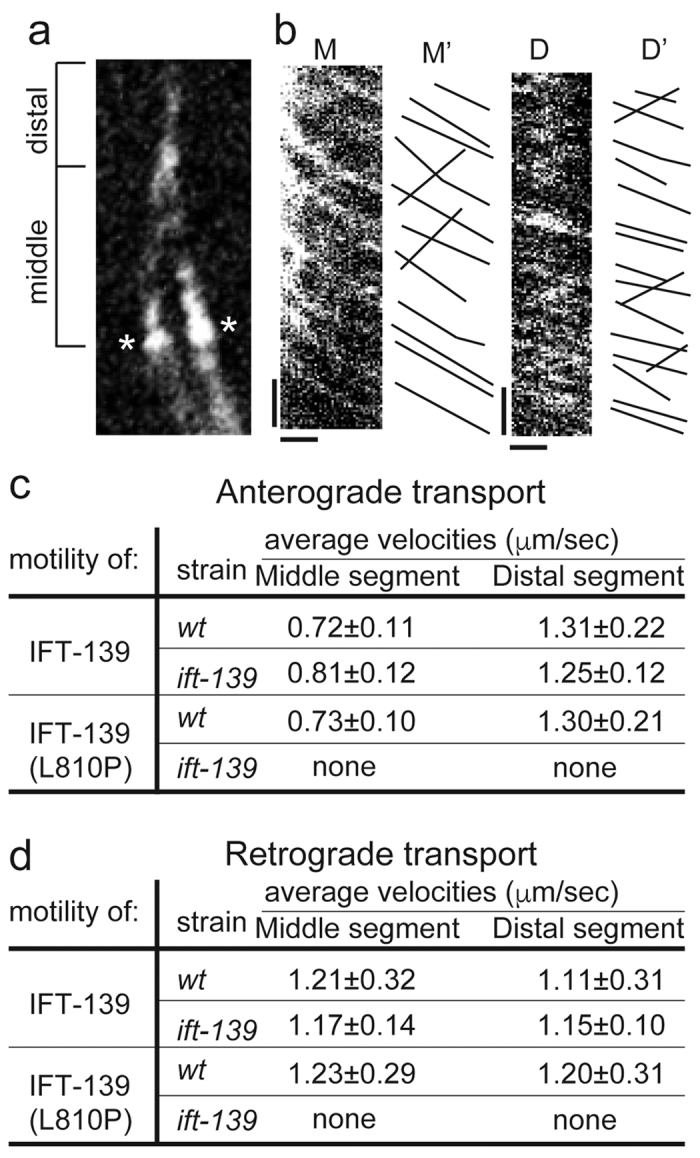
(a) Middle segment and distal segment of cilia are shown. *Indicates the transition zone. (b) Kymographs showing the motility of IFT-139::GFP in phasmid cilia. M and D show kymographs from the middle and distal segments, respectively. M’ and D’ show the trace from M and D. Bars, 5 s (y axis) and 1 μm (x axis). Look at supplemental movie S1. (c) Summary of motility parameters for the anterograde transport. As the ift-139(gk477) mutant expressing IFT-139(L810P) does not have normal cilia (Fig. 4d), the motility cannot be measured. n = 150 particles. Note that IFT-139::GFP moved in wt and ift-139(gk477) in a similar manner. IFT-139(L810P)::GFP moved in wt worms, but cannnot rescue ift-139(gk477). (d) Summary of motility parameters for the retrograde transport. n = 60 particles.
Discussion
The biochemistry of IFT is well studied in Chlamydomonas1,10,11, where the IFT complex is composed of at least 16 protein subunits. Most subunits are conserved among eukaryotes with cilia. The IFT139 subunit is important because mutations in TTC21B that encodes human IFT139 lead to ciliopathies such as NPHP12 and short-rib thoracic dysplasia 423,24,27. While previous studies have identified and characterised most IFT subunits in the nematode, whether or not the C. elegans IFT complex contains and requires the IFT139 subunit remained uncertain. In this paper, I identified and characterised nematode ift-139 and showed that the IFT139 subunit is essential for the formation of normal cilia. The database predicted that ift-139 gene encodes two isoforms. However, only the long isoform (isoform a), but not the short isoform (isoform b), was essential for the intraflagellar transport (Fig. 2d–f). One possibility is that the short isoform is a prediction error. It is difficult to confirm the presence of the short isoform because of the gene structure (Fig. 2a). Another possibility is that the short isoform has roles that are different from the intraflagellar transport. Northern blotting to separate two isoforms and 3’-RACE experiments to determine the transcriptional termination would be needed to confirm the presence of the short isoform.
Genetic data suggest that IFT-139 works together with the products of ifta-1 and che-3. ifta-1 and che-3 encode IFT122 and cytoplasmic dynein-2 heavy chain, both proteins that are required for retrograde IFT39,41. Thus, genetic data suggest that IFT-139 is a component of the IFT-A subcomplex in C. elegans and required for retrograde transport. These genetic data are consistent with the biochemical data in Chlamydomonas that shows that IFT139 belongs to IFT-A42. Interestingly, the length of cilia is significantly longer in ift-139 mutants when a second mutation is introduced in klp-11 (Fig. 5g). There are two possible explanations for how the klp-11 mutation recovers cilia length in ift-139 mutants. One possibility is that the short and bulged cilia are a result of a traffic jam induced by the ift-139 mutation. As klp-11 is an anterograde motor protein, loss of klp-11 reduces the amount of anterograde transport, leading to the reduction of the traffic jam. This is supported by the observation that the amount of OSM-6::GFP is reduced in klp-11 cilia38 (Fig. 5c). This idea is also consistent with the genetic data suggesting that IFT-139 is involved in retrograde transport. While it is not exclusive, another possibility is that KLP-11 transports some factors that negatively regulate cilia length43,44. This hypothesis is supported by the observation that klp-11 mutant males have longer male-specific cilia45.
Disease-associated mutations often give valuable insights into the function of proteins. IFT-139(L810P) is the corresponding mutation to TTC21B(L795P) that causes ciliopathy. Previous studies in zebrafish have showed that TTC21B(L795P) is normally localised to the base of cilia23. We could observe the motility of this mutant protein in C. elegans and found that the IFT-139(L810P) protein is incorporated into the IFT particles and moves normally and bidirectionally in cilia, but cannot rescue the ift-139 mutant (Fig. 6). Together, these results suggest that this residue is not required for the binding of IFT139 to the IFT complex, but the activity of IFT139 is disrupted by this mutation. It is possible that this residue is required to bind to a more peripheral subunit, or IFT cargo molecules such as cilia tubulin46. Further investigations are required to fully analyse and test these molecular interactions; however, biochemical analysis of IFT has not been established in C. elegans. Nevertheless, our genetic data will inform future biochemical studies in Chlamydomonas and mammals to reveal both how IFT139 is incorporated into, and its role in, the IFT complex.
Methods
Strains and Genetics
Caenorhabditis elegans was cultured and maintained according to the standard protocol12. Genotypes were determined by PCR and genomic sequencing in deletion mutants and point mutations, respectively. Primer sets used to detect mutants were as follows: ift-139(gk477): 5′-GGCAGTAGCATACGAAGTGTAAGGAG-3′, 5′-CTCTCCAGTGCTCCACATCCTTGTG-3′ and 5′-GCTGTATACACCACCTAAAGCCTACC-3′. klp-11(tm324): 5′-GCTCACACATTGACATAGGCCGTCG-3′, 5′-GAGACACCACTATCGGCACCAGATG-3′ and 5′-GGCTTCTTATTGTTGAAAACTTCATCCCTCG-3′. ifta-1(gk1004): 5′-GCACTGCTTCAACTCTTCATATGGG-3′, 5′-CCCTTCATCATCGTCATCATTTCGAATC-3′ and 5′-AGTGCCGTGCGGAACCCAGTAGATG-3′. che-3: 5′-TTTGCTTGGACTTGCTTGGAGCTTG-3′ and 5′-GTACTTCTGTTCAATGGATTCTTGCCTACC-3′ for PCR and 5′-TCATATTTGGGCGATCCTTTGGTCTC-3′ for sequencing. Strains are described in Supplemental Table S1.
Construction of ift-139 promoter::gfp and cloning of ift-139 cDNA
The ift-139 promoter, defined as the region between the last exon of the next upstream gene and the start codon of ift-139, was amplified from genomic DNA extracted from wild-type worms Primer sets: 5′-CTGCATGCTCCATACTCGAATACTGAG-3′ and 5′-atGGCGCGCCAAATTGAGATATCAGCAATAAAAATAT-3′. SphI and AscI sites were added to ligate the PCR product to the pSM vector. A C. elegans cDNA library was a generous gift from K. Mizumoto47. ift-139 cDNA was amplified from the library by 2 step. Firstly, PCR was performed using 5′ UTR and 3′ UTR primer pairs and then second PCR was performed and cloned into the pSM vector using NheI and KpnI sites. PCR primers were as follows: PCR #1 was 5′-GTCAAAGCAATATTTTTATTGCTGATATCTC5′-TCACAAATGAAAGTATTCCCCATTTAC-3′ and -3′. PCR #2 was 5′-atGCTAGCaaaaATGGATTCCGAATCTGACGATAATCCAAATG-3′ and 5′-atgcGGTACCcc AGTTCTGATTAGAGCTTTCGCTTTATCCAT-3′.
Chemotaxis assay
Well-fed adult worms were washed four times with phosphate-buffered saline. A 10-cm Petri dish containing a standard assay surface (2% agar, 5 mM K2PO4, 1 mM CaCl2 and 1 mM MgSO4) was used for the assay. Two X marks 180° opposite each other were made near the edge of the plate. 1M NaN3 was absorbed to these two points. After allowing the NaN3 to dry, approximately 50 worms were pipetted onto the plate. Quickly, 1/100 diluted diacetyl or ethanol were placed on each X mark and the lid was closed. After 1 hour, the number of worms was counted and the chemotaxis index was calculated as follows: (number of worms in diacetyl zone − number of worms in control zone)/(total number of worms).
RT-PCR assay
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific, MA, USA). cDNA was synthesized using SuperScript IV reverse transcriptase (Thermo Fisher Scientific) according to the manufacture’s protocol. PCR was performed using KOD FX neo DNA polymerase (TOYOBO, Tokyo, JAPAN). Primer sets are following: ift-139: 5′-CTCCATTTGAGCGAGATCCTGAGC-3′ and 5′-CAGCCAGAATTGAGTTAGCTTCATCG-3′. tba-1: 5′-GTACACTCCACTGATCTCTGCTGACAAG-3′ and 5′-CTCTGTACAAGAGGCAAACAGCCATG-3′48.
Male mating assay
Males were produced by heat shock procedure. 10–15 plates that have 10 L4 worms were placed in 30 °C for 8 hours and then transferred to 20 °C. 4 days later, males were picked up. Typically, more than 20 males were obtained from all genotypes. For mating assay, 3 wt hermaphrodites were mixed with 15 males that were labelled by mnIs17. Mating success was judged by the presence of mnIs17 in the F1 generation. When no F1 animals with the mnIs17 marker was found, I considered the strain exhibited defects in male mating. The phenotype was confirmed by repeating the same procedure.
dauer-formation assay
Plates were starved for 1 week and then worms were observed using a Stemi 508 stereo microscope (Carl Zeiss, Jena, Germany). Three independent plates were prepared for each strains. Dauer was judged by the lack of pharyngeal pumping. When I could not find any dauer worms from 3 independent plates, I considered the strain dauer defective (daf). When the strain is judged as daf in the first assay, the phenotype was confirmed by repeating the same procedure.
dye-filling assay
DiO (Thermo Fisher Scientific) was dissolved in DMSO (Sigma) at 2 mg/ml and stocked at −30 °C. Well-fed worms were collected from a 60-mm plate with 1 ml M9 buffer and pelleted at 1400 g for 2 min. Worms were resuspended in 1 ml M9 buffer and DiO was added (final 0.01 mg/ml). The mixture was slowly shaked for 3 hours. Worms were washed with M9 buffer twice. Fluorescent signal was observed with an Axiovert A1 fluorescent microscope (Carl Zeiss).
Visualisation of cilia structure
Cilia were visualised using mnIs17 expressing osm-6::gfp. OSM-6::GFP is concentrated in cilia and the base of cilia. Phasmid PHA/PHB cilia were mainly observed because the structure was easily and reproducibly detected. Amphid cilia were clouded and seemed different depending on the body angle. They were not analysed in detail. An Axiovert microscope (Carl Zeiss, Jena, Germany) equipped with a LSM710 confocal system (Carl Zeiss) and a Plan Apochromat (x63, NA1.4) objective was used to detect fluorescent signals.
Visualisation of the IFT
Worms expressing IFT-139::GFP or IFT-139(P810L)::GFP were mounted on agarose pads and immobilised with 2 mM levamisole. Phasmid cilia were analysed on an IX81 microscope (Olympus, Tokyo, Japan) equipped with a 100× objective lens (NA1.35) and a CSU-X1 spinning disk confocal head (Yokogawa, Tokyo, Japan). The frame rate was 4 frames/s. Kymographs were made using Image J (National Institutes of Health) using the Multiple Kymograph plugin.
Additional Information
How to cite this article: Niwa, S. The nephronophthisis-related gene ift-139 is required for ciliogenesis in Caenorhabditis elegans. Sci. Rep. 6, 31544; doi: 10.1038/srep31544 (2016).
Supplementary Material
Acknowledgments
I thank Dr. Asako Sugimoto (Tohoku Univ.) and members of the Sugimoto laboratory for helpful discussions and Dr. Mizumoto Kota (British Columbia Univ.) for providing worm cDNA. Some strains were provided by the C.elegans Genetic Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work is supported by the Japan Society for the Promotion of Science KAKENHI Grant No. 15H06033 and the Tomizawa Jun-ichi and Keiko Fund of Molecular Biology Society of Japan for Young Scientists.
Footnotes
Author Contributions S.N. conceived the work, performed all the experiments, prepared all the figures and wrote the paper.
References
- Rosenbaum J. L. & Witman G. B. Intraflagellar transport. Nat Rev Mol Cell Biol 3, 813–825, doi: 10.1038/nrm952 (2002). [DOI] [PubMed] [Google Scholar]
- Ishikawa H. & Marshall W. F. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol 12, 222–234, doi: 10.1038/nrm3085 (2011). [DOI] [PubMed] [Google Scholar]
- Scholey J. M. Cilium assembly: delivery of tubulin by kinesin-2-powered trains. Curr Biol 23, R956–R959, doi: 10.1016/j.cub.2013.09.032 (2013). [DOI] [PubMed] [Google Scholar]
- Jin H. et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219, doi: 10.1016/j.cell.2010.05.015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M. V. et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213, doi: 10.1016/j.cell.2007.03.053 (2007). [DOI] [PubMed] [Google Scholar]
- Ansley S. J. et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425, 628–633, doi: 10.1038/nature02030 (2003). [DOI] [PubMed] [Google Scholar]
- Nonaka S. et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837 (1998). [DOI] [PubMed] [Google Scholar]
- Mochizuki T. et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272, 1339–1342 (1996). [DOI] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol 219, 115–155 (2002). [DOI] [PubMed] [Google Scholar]
- Cole D. G. et al. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141, 993–1008 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Beech P. L. & Rosenbaum J. L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol 131, 1517–1527 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F. & Zhou W. Nephronophthisis-associated ciliopathies. J Am Soc Nephrol 18, 1855–1871, doi: 10.1681/ASN.2006121344 (2007). [DOI] [PubMed] [Google Scholar]
- Cevik S. et al. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol 188, 953–969, doi: 10.1083/jcb.200908133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. et al. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am J Hum Genet 89, 713–730, doi: 10.1016/j.ajhg.2011.11.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambacher N. J. et al. TMEM107 recruits ciliopathy proteins to subdomains of the ciliary transition zone and causes Joubert syndrome. Nat Cell Biol 18, 122–131, doi: 10.1038/ncb3273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wei Q., Zhang Y., Ling K. & Hu J. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol 189, 1039–1051, doi: 10.1083/jcb.200912001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton-Pitt S. R. et al. Ciliogenesis in Caenorhabditis elegans requires genetic interactions between ciliary middle segment localized NPHP-2 (inversin) and transition zone-associated proteins. J Cell Sci 125, 2592–2603, doi: 10.1242/jcs.095539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M., Ishihara T. & Katsura I. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development 126, 4839–4848 (1999). [DOI] [PubMed] [Google Scholar]
- Efimenko E. et al. Caenorhabditis elegans DYF-2, an orthologue of human WDR19, is a component of the intraflagellar transport machinery in sensory cilia. Mol Biol Cell 17, 4801–4811, doi: 10.1091/mbc.E06-04-0260 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales P. L. et al. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat Genet 39, 727–729, doi: 10.1038/ng2038 (2007). [DOI] [PubMed] [Google Scholar]
- Bredrup C. et al. Ciliopathies with skeletal anomalies and renal insufficiency due to mutations in the IFT-A gene WDR19. Am J Hum Genet 89, 634–643, doi: 10.1016/j.ajhg.2011.10.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E. E. et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 43, 189–196, doi: 10.1038/ng.756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh Cong E. et al. A homozygous missense mutation in the ciliary gene TTC21B causes familial FSGS. J Am Soc Nephrol 25, 2435–2443, doi: 10.1681/ASN.2013101126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D. G. & Snell W. J. SnapShot: Intraflagellar transport. Cell 137, 784–784.e781, doi: 10.1016/j.cell.2009.04.053 (2009). [DOI] [PubMed] [Google Scholar]
- Blacque O. E. et al. Functional genomics of the cilium, a sensory organelle. Curr Biol 15, 935–941, doi: 10.1016/j.cub.2005.04.059 (2005). [DOI] [PubMed] [Google Scholar]
- Tran P. V. et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet 40, 403–410, doi: 10.1038/ng.105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C., Li L., Esparza J. M. & Dutcher S. K. Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics 183, 885–896, doi: 10.1534/genetics.109.101915 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T. & Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43, D257–D260, doi: 10.1093/nar/gku949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J., Spike C. A., Lundquist E. A., Shaw J. E. & Herman R. K. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148, 187–200 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Gengyo-Ando K., Ishihara T., Katsura I. & Mitani S. IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells 12, 593–602, doi: 10.1111/j.1365-2443.2007.01076.x (2007). [DOI] [PubMed] [Google Scholar]
- De Riso L., Ristoratore F., Sebastiano M. & Bazzicalupo P. Amphid defective mutant of Caenorhabditis elegans. Genetica 94, 195–202 (1994). [DOI] [PubMed] [Google Scholar]
- Vowels J. J. & Thomas J. H. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130, 105–123 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr M. M. & Sternberg P. W. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature 401, 386–389, doi: 10.1038/43913 (1999). [DOI] [PubMed] [Google Scholar]
- Sengupta P., Chou J. H. & Bargmann C. I. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84, 899–909 (1996). [DOI] [PubMed] [Google Scholar]
- Ward S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci USA 70, 817–821 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Blacque O. E., Snow J. J., Leroux M. R. & Scholey J. M. Functional coordination of intraflagellar transport motors. Nature 436, 583–587, doi: 10.1038/nature03818 (2005). [DOI] [PubMed] [Google Scholar]
- Snow J. J. et al. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol 6, 1109–1113, doi: 10.1038/ncb1186 (2004). [DOI] [PubMed] [Google Scholar]
- Wicks S. R., de Vries C. J., van Luenen H. G. & Plasterk R. H. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol 221, 295–307, doi: 10.1006/dbio.2000.9686 (2000). [DOI] [PubMed] [Google Scholar]
- Hao L., Efimenko E., Swoboda P. & Scholey J. M. The retrograde IFT machinery of C. elegans cilia: two IFT dynein complexes? PLoS One 6, e20995, doi: 10.1371/journal.pone.0020995 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E. et al. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol Biol Cell 17, 5053–5062, doi: 10.1091/mbc.E06-06-0571 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal R. H. et al. Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. J Biol Chem 287, 11689–11703, doi: 10.1074/jbc.M111.287102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S. et al. KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev Cell 23, 1167–1175, doi: 10.1016/j.devcel.2012.10.016 (2012). [DOI] [PubMed] [Google Scholar]
- He M. et al. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol 16, 663–672, doi: 10.1038/ncb2988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsci N. S. & Barr M. M. Kinesin-3 KLP-6 regulates intraflagellar transport in male-specific cilia of Caenorhabditis elegans. Curr Biol 21, 1239–1244, doi: 10.1016/j.cub.2011.06.027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L. et al. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol 13, 790–798, doi: 10.1038/ncb2268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K. & Shen K. Interaxonal interaction defines tiled presynaptic innervation in C. elegans. Neuron 77, 655–666, doi: 10.1016/j.neuron.2012.12.031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J. & Vanfleteren J. R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9, 9, doi: 10.1186/1471-2199-9-9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.