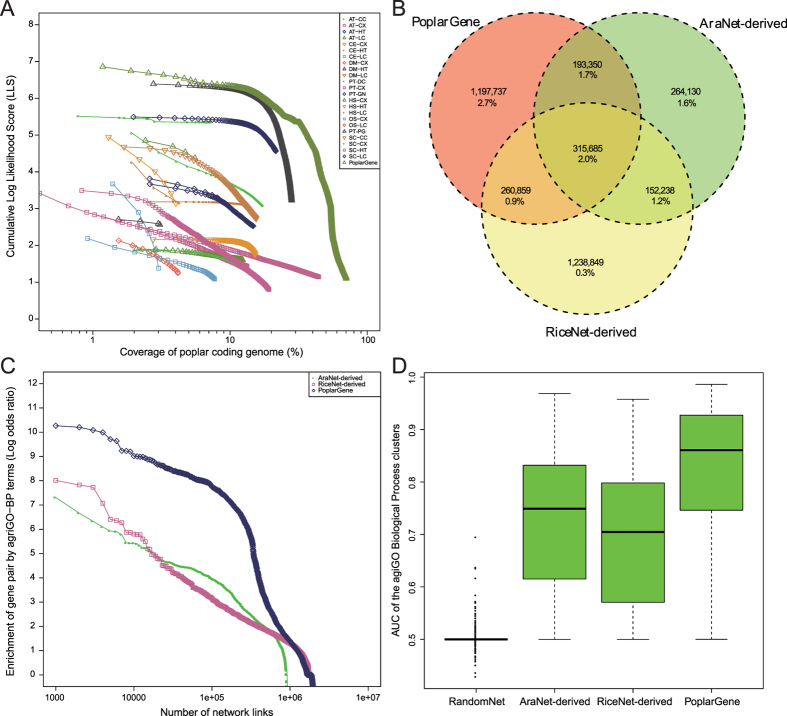
Figure 2. Summary of quality assessment of the PoplarGene network.
(A) The gene linkages derived from 23 diverse functional genomics data sets, representing millions of experimental or computational observations, were integrated into a comprehensive network with higher accuracy and genome coverage than any single data set. The integrated network contains 1,967,631 linkages and 29,049 genes (>70% of the P. trichocarpa coding genome). The x-axis represents the log-scaled coverage of the P. trichocarpa coding genome covered by linkages derived from the corresponding datasets (curves). The y-axis indicates the accuracy of functional linkages, measured as the cumulative log likelihood of linked genes to shared GO-BP term annotations tested using 0.632 bootstrapping and plotted for each bin of 1,000 linkages. The datasets were designated AA-BB, with AA indicating species of data origin (AT, A. thaliana; CE, C. elegans; DM, D. melanogaster; HS, H. sapiens; OS, O. sativa; PT, P. trichocarpa; SC, S. cerevisiae) and BB indicating data type (CC, co-citation; CX, mRNA coexpression; DC, domain co-occurrence; GN, gene neighbor; LC, literature curated protein interactions; HT, high-throughput experimental screening of interaction; PG, phylogenetic profiles). (B) Venn diagram of the gene linkages, indicating that the PoplarGene network contains many more linkages than those derived by orthology transfer from the Arabidopsis gene network AraNet12 and the rice gene network RiceNet32 and that they have higher linkage accuracy. Linkage accuracy was measured using an independent set of reference linkages obtained from the agriGO database. (C) Precision-recall analysis comparing the PoplarGene network to the AraNet-derived network and the RiceNet-derived network. (D) Box-and-whisker plot of network predictive power for 277 agriGO BP terms (with more than four annotated genes), as measured by the area under the curve from ROC analysis.